Clinical Knowledge Graph Construction and Evaluation with Multi-LLMs via Retrieval-Augmented Generation
📝 Original Paper Info
- Title: Clinical Knowledge Graph Construction and Evaluation with Multi-LLMs via Retrieval-Augmented Generation- ArXiv ID: 2601.01844
- Date: 2026-01-05
- Authors: Udiptaman Das, Krishnasai B. Atmakuri, Duy Ho, Chi Lee, Yugyung Lee
📝 Abstract
Large language models (LLMs) offer new opportunities for constructing knowledge graphs (KGs) from unstructured clinical narratives. However, existing approaches often rely on structured inputs and lack robust validation of factual accuracy and semantic consistency, limitations that are especially problematic in oncology. We introduce an end-to-end framework for clinical KG construction and evaluation directly from free text using multi-agent prompting and a schema-constrained Retrieval-Augmented Generation (KG-RAG) strategy. Our pipeline integrates (1) prompt-driven entity, attribute, and relation extraction; (2) entropy-based uncertainty scoring; (3) ontology-aligned RDF/OWL schema generation; and (4) multi-LLM consensus validation for hallucination detection and semantic refinement. Beyond static graph construction, the framework supports continuous refinement and self-supervised evaluation, enabling iterative improvement of graph quality. Applied to two oncology cohorts (PDAC and BRCA), our method produces interpretable, SPARQL-compatible, and clinically grounded knowledge graphs without relying on gold-standard annotations. Experimental results demonstrate consistent gains in precision, relevance, and ontology compliance over baseline methods.💡 Summary & Analysis
1. **First Contribution**: Our first contribution is the KG-RAG framework that integrates multi-agent prompting, LLM-based refinement, and continuous evaluation without relying on golden-standard labels. This can be compared to a team of experts working together to design and review a complex building. 2. **Second Contribution**: We introduce a schema-constrained, FHIR-aligned EAV extraction module paired with ontology mapping and OWL-based encoding. Much like using various tools to create precise architectural blueprints, this allows us to structure complex medical information effectively. 3. **Third Contribution**: Our method empirically validates the system on oncology narratives using metrics that span factuality, semantic grounding, and relational completeness. This is akin to a doctor analyzing complex patient data to make an accurate diagnosis.📄 Full Paper Content (ArXiv Source)
Constructing accurate and clinically relevant knowledge graphs (KGs) from unstructured medical narratives is a foundational challenge in biomedical informatics. Clinical KGs enable explainable AI, decision support, and longitudinal patient modeling, yet traditional approaches remain limited. Rigid schemas like FHIR often lack semantic flexibility , and while ontologies such as SNOMED CT , LOINC , and RxNorm offer standard terminologies, they struggle to capture the temporal, contextual, and inferential nuances needed for oncology and other complex domains.
Conventional rule-based or manual KG construction pipelines are brittle, difficult to scale, and ill-suited to the evolving language and structure of clinical narratives. In contrast, large language models (LLMs) have emerged as powerful tools for semantic parsing, relation discovery, and context-aware generation. Models such as Gemini 2.0 Flash , GPT-4o , and Grok 3 show promise for automating KG construction directly from text. However, their outputs are prone to hallucinations, semantic drift, and factual inconsistency—issues especially critical in high-stakes domains like oncology .
Recent works such as CancerKG.ORG , EMERGE , and CLR2G explore LLM integration with structured or multimodal sources, but lack generalizable pipelines for KG construction and validation directly from clinical text.
This paper introduces the first end-to-end framework for constructing and evaluating clinical knowledge graphs from free-text using multi-agent prompting and a graph-based Retrieval-Augmented Generation (KG-RAG) approach. Our pipeline supports continuous refinement and self-supervised evaluation, enabling both high-precision construction and dynamic graph improvement over time.
We leverage a multi-agent LLM pipeline combining the complementary strengths of:
-
Gemini 2.0 Flash for schema-guided Entity–Attribute–Value (EAV) extraction;
-
GPT-4o for contextual enrichment, ontology alignment, and reflection-based refinement;
-
Grok 3 for validation through contradiction testing and conservative filtering.
All triples are mapped to SNOMED CT, LOINC, RxNorm, GO, and ICD, and encoded in RDF/RDFS/OWL for semantic reasoning. Trust metrics are derived from model agreement and alignment with biomedical ontologies. We demonstrate our method on 40 clinical oncology reports from the CORAL dataset , spanning PDAC and BRCA cohorts, and evaluate triple correctness, ontology coverage, relation diversity, and graph connectivity.
Our key contributions are as follows.
-
We propose a KG-RAG framework that integrates multi-agent prompting, LLM-based refinement, and continuous evaluation without gold-standard labels.
-
We introduce a schema-constrained, FHIR-aligned EAV extraction module paired with ontology mapping and OWL-based encoding.
-
We support inference over implicit, cross-sentence, and multi-attribute relations with robust semantic validation.
-
We encode the final graphs with semantic web standards and introduce composite trust scoring mechanisms.
-
We empirically validate our system on oncology narratives using metrics spanning factuality, semantic grounding, and relational completeness.
By uniting multi-LLM consensus, ontology grounding, and iterative refinement, our system offers a scalable, explainable, and verifiable approach for clinical KG construction—laying the foundation for real-time decision support and next-generation clinical AI.
Related Work
Recent advancements in hybrid knowledge graph–language model (KG–LLM) systems, retrieval-augmented generation (RAG), and contrastive learning have significantly influenced clinical informatics. However, many existing approaches rely on predefined schemas, curated corpora, or multimodal data. Our work addresses a critical gap: enabling schema-inductive, verifiable knowledge graph construction directly from unstructured clinical narratives.
Schema-Constrained KG–LLM Hybrids
CancerKG.ORG integrates large language models (LLMs) with curated colorectal cancer knowledge using models such as GPT-4, LLaMA-2, and FLAN-T5, alongside structured meta-profiles and guardrailed RAG. Other frameworks like KnowGL , KnowGPT , and KALA demonstrate the efficacy of knowledge graph alignment but depend on static ontologies. In contrast, our system facilitates dynamic schema evolution through automatic entity–attribute–value (EAV) extraction, probabilistic triple scoring, and iterative ontology grounding—eliminating the need for manual curation or fixed schemas.
Multimodal RAG and Predictive Systems
Systems such as EMERGE , MedRAG , and GatorTron-RAG combine clinical notes, structured electronic health record (EHR) data, and biomedical graphs for predictive tasks. While effective for diagnosis or risk stratification, these models focus on classification rather than symbolic knowledge representation. Our approach emphasizes interpretable, symbolic triple construction using a fully text-based pipeline—removing dependencies on external modalities or fixed task objectives.
Contrastive and Trust-Aware LLM Generation
Trust-aware LLM generation has been explored through methods like Reflexion and TruthfulQA , which employ self-reflection and hallucination mitigation techniques. Similarly, CLR2G applies cross-modal contrastive learning for radiology report generation. While these approaches enhance semantic control, our framework advances this by introducing multi-agent triple scoring, entropy-aware trust filtering, and reflection-based validation across LLMs such as Gemini, GPT-4o, and Grok—focusing specifically on symbolic reasoning from clinical text.
Structured Retrieval and Table-Centric Knowledge Graphs
Earlier platforms including WebLens , Hybrid.JSON , and COVIDKG introduced scalable structural retrieval and metadata modeling for heterogeneous clinical data tables. CancerKG extends this direction using vertical and horizontal metadata modeling over structured cancer databases. Our work fundamentally differs in modality and scope: rather than relying on table alignment or attribute normalization, we construct evolving clinical knowledge graphs directly from free-text reports using prompt-based decomposition, probabilistic scoring, and ontology-backed reasoning.
In summary, while existing systems have laid essential groundwork in structured knowledge extraction and LLM-KG integration, our contribution is a self-evaluating, schema-flexible pipeline that autonomously discovers, validates, and encodes clinical knowledge from raw narratives. By removing rigid schema dependencies and leveraging multi-LLM agreement for factual trust, we enable scalable, interpretable, and clinically relevant knowledge graph construction that dynamically adapts to new domains and document types.
Methodology
Overview of Multi-Agent KG Construction and Evaluation Pipeline
We propose a multi-agent framework for constructing clinically verifiable and semantically interoperable knowledge graphs (KGs) directly from oncology narratives. The system orchestrates three state-of-the-art large language models—Gemini 2.0 Flash, GPT-4.o, and Grok 3—each assigned a specialized role across five modular stages. This architecture is designed to produce structured, explainable, and ontology-aligned knowledge graphs from unstructured clinical text.
-
EAV Extraction (Gemini 2.0 Flash): Using tailored prompts and FHIR-aware templates, Gemini 2.0 Flash performs extraction of Entity–Attribute–Value (EAV) triples from free-text narratives. Extracted entities are linked to canonical clinical resource types.
-
Ontology Mapping (Gemini 2.0 Flash): Attributes and values are mapped to standard biomedical vocabularies such as SNOMED CT, LOINC, and RxNorm. This step normalizes terminology and facilitates semantic consistency across clinical concepts.
-
Relation Discovery (Gemini 2.0 Flash + GPT-4.o): Semantic relationships between entities and attributes are identified through prompt-driven extraction and refinement. Gemini 2.0 Flash generates candidate links, while GPT-4.o refines and validates relation types using contextual understanding and ontology alignment.
-
Semantic Web Encoding (RDF/RDFS/OWL): All triples are encoded using Semantic Web standards such as RDF, RDFS, and OWL. This enables symbolic reasoning, knowledge graph querying via SPARQL, and integration with external ontology-based systems.
-
KG Validation (Gemini 2.0 Flash + GPT-4.o + Grok 3): A composite trust function is applied to evaluate the reliability of each triple. The trust score incorporates:
-
Self-consistency from Gemini 2.0 Flash (via re-prompting),
-
Semantic grounding from GPT-4.o (evidence retrieval and consistency checks),
-
Robustness verification from Grok 3 (counterfactual and adversarial assessment).
-
This pipeline supports schema-flexible, ontology-informed, and model-validated knowledge graph construction from raw clinical narratives. The modular architecture facilitates downstream reasoning, querying, and integration into clinical decision support systems while maintaining traceability and explainability of the extracted information.
Stage 1: FHIR-Guided EAV Extraction
The first stage of our pipeline focuses on extracting structured clinical knowledge in the form of Entity–Attribute–Value (EAV) triples. This is achieved using schema-guided prompting with Gemini 2.0 Flash, integrating syntactic cues from the narrative and semantic constraints from the FHIR (Fast Healthcare Interoperability Resources) specification. Entities are concurrently typed using FHIR to ensure semantic consistency and interoperability.
Triple Extraction.
Let $`\mathcal{D} = \{x_1, x_2, \ldots, x_N\}`$ be the corpus of clinical narratives. For each $`x_i \in \mathcal{D}`$, we construct a structured prompt $`\pi(x_i)`$ tailored for Gemini 2.0 Flash, which generates a candidate set of EAV triples as:
\mathcal{T}_i = f_{\theta}(\pi(x_i)) = \left\{(e_j, a_j, v_j)\right\}_{j=1}^{k}, \quad \forall i \in [1, N]Each triple $`(e_j, a_j, v_j)`$ includes:
$`e_j \in \mathcal{E}_{\text{FHIR}}`$: a FHIR-typed entity (e.g.,
Procedure, Observation), $`a_j`$: a clinical attribute, and $`v_j`$:
a narrative-grounded value.
FHIR Typing Function.
Entities are normalized via a typing function $`\phi_{\text{FHIR}} : \mathcal{E} \rightarrow \mathcal{E}_{\text{FHIR}}`$, ensuring alignment to standard resource types for downstream mapping.
Entropy-Based Value Confidence.
To evaluate confidence in value predictions, we compute token-level entropy for $`v_j`$ given its sub-token distribution $`P(v_j) = \{p_1, \ldots, p_m\}`$:
H(v_j) = -\sum_{t=1}^{m} p_t \log p_tValues with $`H(v_j) > \delta`$ (threshold $`\delta`$) are flagged for further validation or multi-model filtering.
Illustrative EAV Triples.
Examples include:
(Procedure, performed_by, SurgicalOncologist)
(Observation, hasLabResult, CA 19-9)
(HER2 Status, determines, Trastuzumab Eligibility)
These EAVs serve as the foundation for ontology mapping, relation discovery, and semantic web encoding in the subsequent stages.
Stage 2: Ontology Mapping & Schema Construction
To enable semantic reasoning and interoperability, extracted EAV concepts are mapped to standardized biomedical ontologies via LLM-guided retrieval and similarity alignment. Gemini 2.0 Flash orchestrates this step and produces OWL/RDFS-compliant schemas for graph construction.
Ontology Vocabulary.
We define the ontology set:
\mathcal{O} = \{\text{SNOMED CT}, \text{LOINC}, \text{RxNorm}, \text{ICD}, \text{GO}\}Each $`\mathcal{O}_i`$ contributes domain-specific concepts, e.g.,
SNOMED: Weight Loss, LOINC: CA 19-9, RxNorm: FOLFIRINOX.
Concept Mapping.
Given raw terms $`\mathcal{C}_{\text{raw}} = \{c_1, \ldots, c_M\}`$, define a mapping:
\mu: \mathcal{C}_{\text{raw}} \rightarrow \mathcal{C}_{\text{mapped}} \subseteq \bigcup_i \mathcal{O}_iScoring uses lexical and semantic similarity:
\text{Score}(c_i, o_j) = \alpha \cdot \text{sim}_{\text{lex}} + \beta \cdot \text{sim}_{\text{sem}}, \quad \alpha + \beta = 1Schema Construction.
Mapped concepts are encoded in OWL/RDFS:
-
Class Typing: If $`o_j \in \mathcal{O}_{\text{SNOMED}}`$, declare $`o_j`$ as an OWL
Class. -
Property Semantics:
MATH\begin{align*} \texttt{hasLabResult} &\sqsubseteq \texttt{ObjectProperty}, \\ \texttt{domain(hasLabResult)} &= \texttt{Observation}, \\ \texttt{range(hasLabResult)} &= \texttt{LabTest} \end{align*}Click to expand and view more -
TBox Inclusion:
ElevatedCA19_9$`\sqsubseteq`$AbnormalTumorMarker.
Persistent URIs.
Each concept receives a resolvable URI, e.g.,
http://snomed.info/id/267036007 for “Weight Loss.”
Outcome.
This stage yields an ontology-aligned schema that supports OWL reasoning, SPARQL queries, and Linked Data integration, while ensuring formal consistency and semantic traceability.
Stage 3: Relation Discovery
To move beyond isolated EAV triples, this stage enriches the knowledge graph with typed relations capturing diagnostic reasoning, temporal dependencies, and treatment logic. Structured prompting and multi-agent validation (Gemini 2.0 Flash, GPT-4.o, Grok 3) are used to discover and filter semantic relations.
Relation Typing. Each relation $`r_i \in \mathcal{R}`$ is classified as:
r_i \in
\begin{cases}
\mathcal{R}_{EE} & \text{(Entity–Entity)} \\
\mathcal{R}_{EA} & \text{(Entity–Attribute)} \\
\mathcal{R}_{AA} & \text{(Attribute–Attribute)}
\end{cases}Examples include: $`\mathcal{R}_{EE}`$:
Biopsy $`\rightarrow`$confirms$`\rightarrow`$ TumorType,
$`\mathcal{R}_{EA}`$:
CT Scan $`\rightarrow`$visualizes$`\rightarrow`$ Pancreatic Mass,
$`\mathcal{R}_{AA}`$:
HER2 Status $`\rightarrow`$determines$`\rightarrow`$ Trastuzumab Eligibility.
Candidate Generation. Given narrative $`x`$, candidate relations $`\mathcal{T}_{\text{rel}} = \{(h_i, p_i, t_i)\}`$ are generated via Gemini:
\mathcal{T}^{\text{gen}}_{\text{rel}} = f_{\text{Gemini}}(\pi_{\text{rel}}(x))with $`h_i, t_i \in \mathcal{E} \cup \mathcal{A}`$ and $`p_i \in \mathcal{V}_{\text{verb}}`$.
Semantic Validation. Each triple $`\tau_i`$ is scored by GPT-4.o using contextual inference:
J(\tau_i) = f_{\text{GPT-4.o}}(\pi_{\text{judge}}(\tau_i, x)) \in [0,1]Adversarial Filtering. Grok 3 perturbs each relation and flags contradictions:
\xi(\tau_i) = \frac{|\{\tau_i' \in \mathcal{A}(\tau_i)\ |\ \texttt{contradictory}\}|}{|\mathcal{A}(\tau_i)|}Final Set. The accepted relation set filters for high plausibility and low contradiction:
\mathcal{T}_{\text{rel}}^{\text{trusted}} = \left\{\tau_i \mid J(\tau_i) > \delta,\ \xi(\tau_i) \leq \epsilon \right\}Outcome. This step yields a validated set of typed relations interlinking FHIR-grounded concepts. These enhance the inferential depth of the KG, supporting diagnostic, prognostic, and treatment-oriented reasoning.
Stage 4: Semantic Graph Encoding
To enable reasoning and system interoperability, validated triples $`\tau_i = (s_i, p_i, o_i)`$ are encoded using Semantic Web standards: RDF, RDFS, OWL, and SWRL.
RDF Triples. Each assertion is modeled as: $`(s_i, p_i, o_i) \in \mathcal{G}_{\text{RDF}}`$, where $`s_i, o_i \in \mathcal{U} \cup \mathcal{L}`$ and $`p_i \in \mathcal{U}`$; $`\mathcal{U}`$ denotes URIs and $`\mathcal{L}`$ literals.
RDFS Constraints. Predicates include domain/range typing: $`\text{domain}(p_i) = C_s`$, $`\text{range}(p_i) = C_o`$, with type assertions: $`\texttt{rdf:type}(s_i, C_s) \land \texttt{rdf:type}(o_i, C_o)`$.
OWL Semantics. Logical rules include: Subclass: $`A \sqsubseteq B`$, Equivalence: $`A \equiv B`$, Restriction: $`\texttt{Biopsy} \sqcap \exists\,\texttt{hasOutcome}.\texttt{Malignant} \sqsubseteq \texttt{PositiveFinding}`$.
SPARQL Query. Query for high Ki-67 index:
PREFIX kg: <http://example.org/kg#>
SELECT ?p WHERE {
?p kg:hasAttribute ?a .
?a rdf:type kg:Ki67_Index .
?a kg:indicates ?v .
FILTER(?v > 20)
}
SWRL Rule. Example rule for identifying high-risk PDAC patients:
\begin{array}{l}
\texttt{Patient}(p) \land \texttt{hasAttribute}(p, ca) \land \texttt{CA19\_9}(ca) \land \\
\quad \texttt{indicates}(ca, v_1) \land \texttt{greaterThan}(v_1, 1000) \land \\
\quad \texttt{hasAttribute}(p, w) \land \texttt{WeightLoss}(w) \land \\
\quad \texttt{indicates}(w, v_2) \land \texttt{greaterThan}(v_2, 10) \\
\Rightarrow \texttt{HighRiskPatient}(p)
\end{array}Outcome. This stage produces an ontology-compliant semantic graph supporting RDFS validation, OWL/SWRL inference, and SPARQL querying—ideal for clinical integration and semantic analytics.
Stage 5: Multi-LLM Trust Validation
| Technique | Formal Rule | Illustrative Example |
|---|---|---|
| Stage 1: Baseline Matching Techniques | ||
| Case-Sensitive Matching | vi ∈ 𝒯exact(dj) | “FOLFIRINOX” matched exactly in raw text |
| Regex Matching | regex("12̇ ?mg/dL") |
bilirubin: 1.2 mg/dL matched
as value |
| Fuzzy String Matching | fuzz.ratio(vi, tk) > τ |
“FOLFRINOX” → “FOLFIRINOX” |
| N-Gram Phrase Matching | ∃ g ⊂ dj: sim(g, vi) > γ | “2–3 episodes/day” matched to vomiting frequency |
| Boolean Inference | “denies smoking” ⇒ smoking: false |
Negation mapped to absent finding |
| Stage 2: Heuristic Augmentation | ||
| Case-Insensitive Matching | lower(v_i) = lower(t_k) |
“Male” == “male” == “MALE” |
| Custom Negation Detection | “denies chills” →
observation_chills: absent |
Handcrafted negation patterns |
| spaCy Lemmatization | lemma(ai) = tk |
“diaphoretic” → “diaphoresis” |
| Synonym Mapping | ai ∈ Σsyn | “dyspneic” ↔︎ “dyspnea” |
| Stage 3: Specialized Resolution | ||
| Sentence-Level Negation | dep(tk) = neg in sentence(vi) | “denies pruritus” →
absent |
| Typo Correction | fuzz.ratio(deneid, denied) > 95 |
Handles transcription errors |
| Explicit Fixes | “smking” → “smoking” | Dictionary-based recovery of high-impact terms |
| Technique | Formal Rule or Criterion | Illustrative Example |
|---|---|---|
| Evidence Alignment (Entailment) | $`\text{Sim}_{\text{entail}}(\tau, e_k)`$ for $`e_k \in \mathcal{E}_\tau`$ | “HER2 is overexpressed” entails HER2 $`\rightarrow`$determines$`\rightarrow`$ Trastuzumab Eligibility |
| LLM Reflective Scoring | $`J(\tau) \in [0, 1]`$ from GPT-4o verifier prompt | GPT-4o returns 0.92 plausibility for therapy eligibility triple |
| Self-Consistency across Prompts | $`C(\tau) = \frac{1}{n} \sum \mathbb{I}[\tau \in \mathcal{T}^{(i)}]`$ | Relation appears in 4/5 prompt variants → $`C = 0.80`$ |
| Domain-Range Schema Check | $`\text{type}(s) \in \text{domain}(p)`$; $`\text{type}(o) \in \text{range}(p)`$ | Tumor → hasMarker → HER2 invalid if HER2 is not range of hasMarker |
| Redundancy Score | $`\cos(\tau_i, \tau_j) > \gamma`$ in embedding space | “confirms” and “verifies” flagged as duplicates |
| Semantic Clustering for Gaps | Use UMAP clustering over embeddings | Reveals that “initiated” and “started” lack edge alignment |
To ensure the reliability and semantic precision of the constructed clinical knowledge graph, we implement a multi-layered validation framework. This framework consists of two complementary tiers: (1) evaluating the fidelity of Entity–Attribute–Value (EAV) triples and (2) verifying semantic relation triples. Both tiers are supported by a multi-agent setup involving Gemini 2.0 Flash, GPT-4.o, and Grok 3, which collaboratively perform grounding verification, reflective reasoning, and adversarial testing.
EAV Validation: Grounding and Fidelity Assessment
Each EAV triple $`(e, a, v)`$ is validated across multiple quality dimensions, including textual grounding, correctness, hallucination risk, recoverability, and prompt-based consistency.
EAV Validation Metrics.
-
Coverage: Whether the attribute $`a`$ and value $`v`$ are directly supported by the source text.
-
Correctness Rate (CR): Proportion of grounded EAV triples verified as correct.
-
Hallucination Rate (HR): Fraction of generated triples lacking explicit textual support.
-
Rescue Rate (RR): Proportion of hallucinated triples that become valid after normalization or lexico-semantic heuristics.
-
Self-Consistency: Agreement across multiple prompting strategies applied to the same source.
Relation Validation: Semantic Integrity and Clinical Utility
Each relation triple $`(s, p, o)`$ is evaluated on criteria that assess its semantic validity, ontological alignment, clinical relevance, and informativeness within the graph.
Relation Evaluation Criteria.
-
Semantic Validity: Verified via entailment and alignment with external evidence or ontologies.
-
Schema Compliance: Ensures each predicate respects RDF domain and range constraints.
-
Clinical Usefulness: Prioritizes relations with diagnostic, prognostic, or therapeutic significance.
-
Structural Role and Frequency: Measures relational salience based on graph centrality and recurrence.
-
Redundancy and Gaps: Detects paraphrased or missing links using embedding-based similarity.
-
Multi-LLM Agreement: Confidence derived from consensus across Gemini 2.0, GPT-4.o, and Grok 3 outputs.
Unified Trust Scoring
A composite trust score $`T(\tau)`$ is assigned to each triple $`\tau = (s, p, o)`$ or $`(e, a, v)`$:
T(\tau) = \lambda_1 R(\tau) + \lambda_2 C(\tau) + \lambda_3 J(\tau), \quad \sum \lambda_i = 1Where:
-
$`R(\tau)`$: Evidence alignment score from entailment or retrieval.
-
$`C(\tau)`$: Self-consistency across prompt variants.
-
$`J(\tau)`$: Reflective plausibility score from model-based judgment.
Only triples satisfying $`T(\tau) \geq \delta_T`$ (e.g., $`\delta_T = 0.65`$) are retained in the final knowledge graph.
Final Graph Validation and Filtering
The final graph $`\mathcal{G}_{\text{final}}`$ is further validated through graph-level filters:
-
Ontology Compliance: Ensures that all relations adhere to predefined schema constraints.
-
Redundancy Elimination: Removes duplicate or semantically equivalent edges.
-
Clinical Coverage: Emphasizes inclusion of medically relevant concepts (e.g., NCCN-aligned markers, therapeutic eligibility).
Outcome.
This multi-agent, multi-criteria validation framework ensures that the final knowledge graph is clinically grounded, semantically coherent, and structurally optimized. The result is a trustworthy KG suitable for deployment in applications such as explainable AI, cohort identification, treatment planning, and clinical decision support.
Results and Evaluation
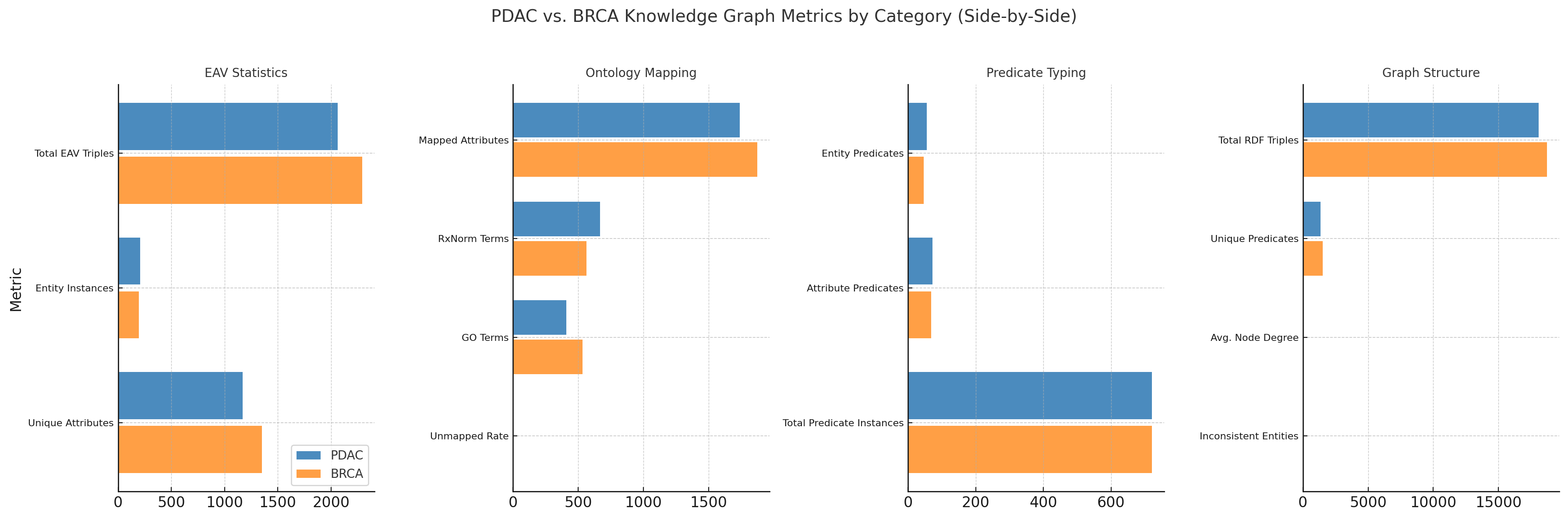
| Metric Category | Metric | PDAC | BRCA | Example / Description |
|---|---|---|---|---|
| EAV Statistics | Total EAV triples | 2,062 | 2,293 | Structured triples such as
(Observation, hasLabResult, CA_19-9). |
| Entity instances | 209 | 195 | Entity types like Procedure,
Observation, Condition. |
|
| Unique attributes | 1,172 | 1,353 | Fine-grained attributes like
Weight_Loss, HER2_Status. |
|
| Ontology Mapping | Mapped attributes | 1,738 | 1,873 | Attributes linked to SNOMED CT, RxNorm, and LOINC. |
| RxNorm terms | 668 | 562 | FOLFIRINOX,
Tamoxifen, chemotherapy agents. |
|
| GO terms | 409 | 533 | Genomic markers like HER2,
BRCA1, EGFR. |
|
| Unmapped rate | 0.69% | 0.85% | Mostly generic strings, typos, or shorthand entries. | |
| Predicate Typing | Entity predicates | 56 | 46 | Action relations:
(Biopsy, confirms, TumorType). |
| Attribute predicates | 72 | 69 | Diagnostic relations:
(HER2Status, determines, TherapyEligibility). |
|
| Total predicate instances | 721 | 721 | Total predicate uses across the graph. | |
| Graph Structure | Total RDF triples | 18,097 | 18,732 | RDF-format atomic facts encoding EAV and relations. |
| Unique predicates | 1,346 | 1,520 | Vocabulary such as
hasLabResult, performedBy. |
|
| Avg. node degree | 5.73 | 5.74 | Graph density and information connectivity. | |
| Inconsistent entities | 25 | 41 | Domain–range issues caught during OWL validation. |
Experimental Setup
We evaluate our clinical knowledge graph (KG) framework using 40 expert-annotated oncology reports from the CORAL (Clinical Oncology Reports to Advance Language Models) dataset. The reports span two cancer types—Pancreatic Ductal Adenocarcinoma (PDAC) and Breast Cancer (BRCA)—which differ significantly in diagnostic focus and treatment documentation.
Our primary objectives are to assess the grounding and accuracy of extracted Entity–Attribute–Value (EAV) triples and to evaluate the structural and semantic integrity of the resulting knowledge graphs. The KG construction pipeline integrates four core components. First, Gemini 2.0 Flash is used to extract FHIR-aligned EAV triples from unstructured narratives. Second, GPT-4.o, Grok 3, and Gemini collaborate to validate triples—filtering hallucinations and resolving inconsistencies. Third, extracted terms are normalized to standardized vocabularies, including SNOMED CT, RxNorm, LOINC, GO, and ICD. Finally, validated triples are encoded into RDF/RDFS/OWL to support SPARQL and SWRL-based rule reasoning.
Each of the 40 clinical reports—20 PDAC and 20 BRCA—contains a narrative
file (.txt), expert annotations (.ann), and model-generated outputs
(.json). The average report is approximately 145,000 tokens, with some
extending to 180,000 tokens in more complex cases. The three LLMs play
distinct but complementary roles. Gemini 2.0 Flash acts as the primary
EAV extractor, aligning outputs to FHIR standards and providing
confidence scores. GPT-4.o validates predicate plausibility and
assigns reflection-based trust levels. Grok 3 performs adversarial
testing to detect redundancy and semantic inconsistencies. This
multi-agent framework enables robust, interpretable, and
ontology-compliant knowledge graph construction directly from raw
clinical narratives.
Knowledge Graph Construction Results
Figure 1 and Table [tab:kg_summary_all] summarize the semantic and structural features of knowledge graphs constructed from PDAC and BRCA oncology reports. The visual and tabular comparisons reflect all key pipeline stages—EAV extraction, ontology mapping, predicate typing, and semantic modeling—and highlight domain-specific characteristics across cancer types. Together, they demonstrate the fidelity, flexibility, and interpretability of our multi-agent LLM-based KG construction approach.
Step 1: FHIR-Aligned EAV Extraction. BRCA records yielded more EAV triples (2,293 vs. 2,062) and a broader attribute set (1,353 vs. 1,172), reflecting molecular precision. Examples:
-
HER2 Status$`\rightarrow`$determines$`\rightarrow`$Trastuzumab Eligibility -
Ki-67 Index$`\rightarrow`$indicates$`\rightarrow`$High Proliferation Rate
PDAC emphasized diagnostic and procedural concepts:
-
CT Scan$`\rightarrow`$visualizes$`\rightarrow`$Pancreatic Mass -
Surgical Resection$`\rightarrow`$treats$`\rightarrow`$Primary Tumor
Step 2: Ontology Mapping. BRCA had more mapped attributes (1,873 vs. 1,738) and GO terms (533 vs. 409), consistent with genomics-rich narratives. PDAC showed stronger RxNorm alignment (668 vs. 562), reflecting treatment-oriented notes. Both cohorts had <1% unmapped rate.
Step 3: Predicate Typing. Both KGs included 721 predicate instances, with PDAC using a broader predicate range (56 entity-level, 72 attribute-level) than BRCA (46, 69). PDAC predicates emphasized procedural workflows, BRCA prioritized stratification and therapeutic relevance.
Step 4: Graph Structure and Semantic Modeling. Both graphs showed dense connectivity (avg. node degree $`\sim`$5.7) and rich vocabularies (PDAC: 1,346 predicates; BRCA: 1,520). OWL validation found more inconsistencies in BRCA (41) than PDAC (25), often from genomics-based domain-range mismatches. Triples were encoded in RDF/RDFS/OWL, supporting SPARQL/SWRL:
\texttt{Biopsy} \sqcap \texttt{hasOutcome.Malignant} \sqsubseteq \texttt{PositiveFinding}Clinical Interpretability. Actionable triples like
(Biopsy, confirms, TumorType) and
(HER2 Status, determines, Trastuzumab Eligibility) demonstrate support
for clinical decision-making and cohort stratification. Our
framework—Gemini 2.0 Flash for generation, GPT-4o for validation, Grok 3
for semantic robustness—transforms unstructured narratives into
queryable, ontology-aligned knowledge graphs.
Evaluation of Knowledge Graphs
We evaluate the quality of LLM-generated clinical knowledge graphs (KGs) using a two-tiered framework: (1) analysis of Entity–Attribute–Value (EAV) triples for textual and semantic fidelity, and (2) structural and relation-level validation of the graph itself. Evaluation spans two cancer cohorts—PDAC and BRCA—derived from the CORAL dataset.
Entity–Attribute–Value (EAV) Evaluation
Evaluation of EAV Extraction Across PDAC and BRCA Cohorts
We conducted a comparative evaluation of entity-attribute-value (EAV) extraction across two cancer cohorts: Pancreatic Ductal Adenocarcinoma (PDAC, Patients 0–19) and Breast Cancer (BRCA, Patients 20–39). Key metrics include raw text and attribute coverage, correctness, hallucinations, and error rates, summarized in Table [tab:eav_summary_detailed].
| Metric | PDAC (P# 0–19) | BRCA (P# 20–39) | ||
|---|---|---|---|---|
| 2-5 | Avg / Rate | Total | Avg / Rate | Total |
| Text & Attribute Coverage | ||||
| Raw Text Coverage (%) | 29.74% | – | 37.97% | – |
| Attribute Coverage (%) | 99.83% | – | 99.83% | – |
| Total Attributes Extracted | – | 2,028 | – | 2,099 |
| Correctness & Error Metrics | ||||
| Correct Attributes (%) | 73.22% | 1,514 | 72.58% | 1,524 |
| Incorrect Attr. (Error#) | 25.45 | 509 | 28.25 | 565 |
| Errors (Attr.) (%) | 25.10% | – | 26.91% | – |
| Hallucination & Consistency | ||||
| Hallucinated Attr. (Total) | 0.20 | 4 | 0.15 | 3 |
BRCA patients showed higher average raw text coverage (37.97%) than PDAC (29.74%), suggesting denser or more structured narratives in breast cancer reports. PDAC achieved slightly better extraction precision, with a lower incorrect attribute rate (25.45 vs. 28.25) and a smaller overall error rate (25.10% vs. 26.91%). Both cohorts demonstrated excellent attribute coverage (99.83%), indicating consistent LLM performance across cancer types. Hallucination rates were minimal—fewer than one per five patients—validating the reliability of prompt-based extraction.
These results highlight a trade-off between recall and precision. BRCA’s richer documentation may increase coverage but introduces more noise, while PDAC yields fewer attributes with higher correctness. This underscores the need for cohort-specific fine-tuning or filtering in future extraction pipelines.
Attribute-Level Comparison
Figure 2 presents a comparison of raw coverage, correctness, and error rates across PDAC and BRCA cohorts. PDAC patients show stable correctness with fewer attribute-level errors, whereas BRCA patients demonstrate greater variability, most notably among Patients 30 and 31, who exhibit high attribute volume but reduced correctness.
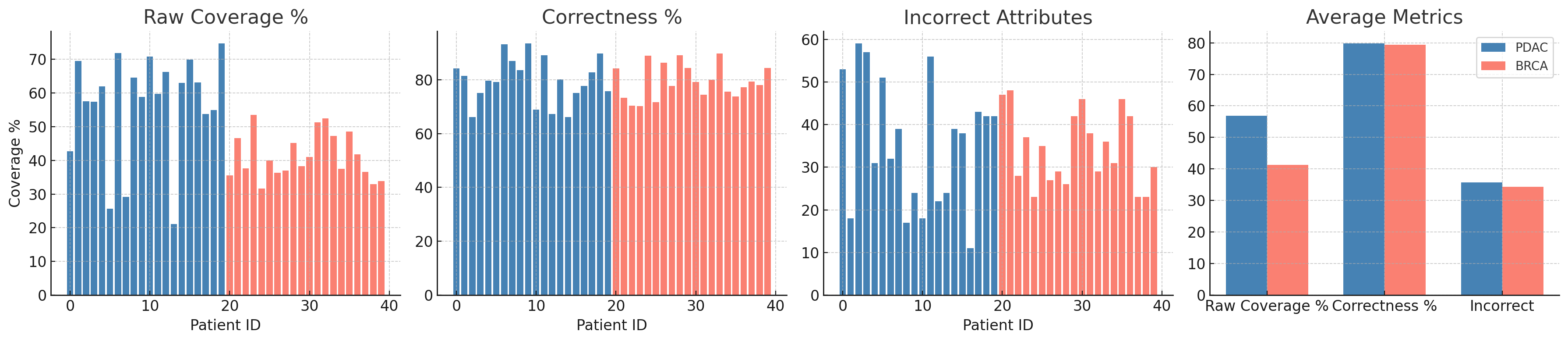
Both cohorts demonstrate strong EAV extraction. PDAC emphasizes higher correctness and reduced noise, while BRCA contributes richer attribute diversity. These patterns highlight the importance of tailoring extraction strategies to cohort-specific narrative styles, especially for oncology KGs and adaptive clinical decision support.
KG Structural and Relation-Level Validation
Multi-LLM Consensus Evaluation of EAV Triples
To ensure semantic fidelity and factual grounding of extracted Entity–Attribute–Value (EAV) triples, we adopt a consensus-driven validation framework using three complementary LLMs: Gemini 2.0 Flash (schema-aware extractor), Grok 3 (evidence-based validator), and GPT-4o (semantic generalizer). Triples are assessed across three dimensions: (1) Factuality—explicit grounding in source text or biomedical ontologies, (2) Plausibility—semantically inferable but implicit triples, and (3) Correction—revision of ambiguous or hallucinated content.
Each model contributes distinct strengths: Grok filters unsupported
relations (e.g., hematuria_etiology $`\rightarrow`$ diagnosis);
GPT-4o proposes contextually inferred alternatives (e.g.,
creatinine $`\rightarrow`$ kidney_function); Gemini provides
precise, schema-aligned outputs.
Figure 3 shows Gemini achieves the highest factual accuracy (98–100%), followed by Grok (94–96%) and GPT-4o (85–92%). Pearson correlations indicate strong alignment between Gemini and Grok ($`r = 0.88`$), with GPT-4o moderately correlated with Gemini ($`r = 0.71`$) and Grok ($`r = 0.69`$), consistent with its broader semantic scope.
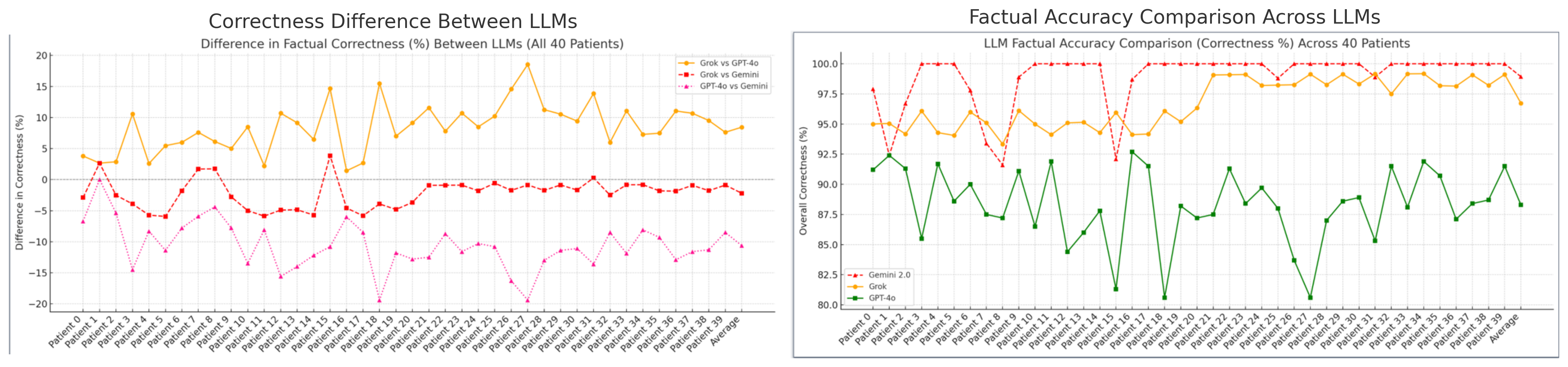
In the absence of gold-standard annotations, we accept triples validated by at least two models. Disagreements are resolved by prioritizing ontology-compliant alternatives (e.g., SNOMED CT, RxNorm), with uncertain triples flagged for reflection and reranking. Together, the precision of Gemini, the filtering strength of Grok, and the contextual breadth of GPT-4o enable robust, unsupervised validation of clinical KGs. Correlation trends reinforce their complementary roles in accurate, ontology-aligned graph construction—without the need for human-labeled supervision.
Statistical Analysis of Relation Diversity and Source Coverage
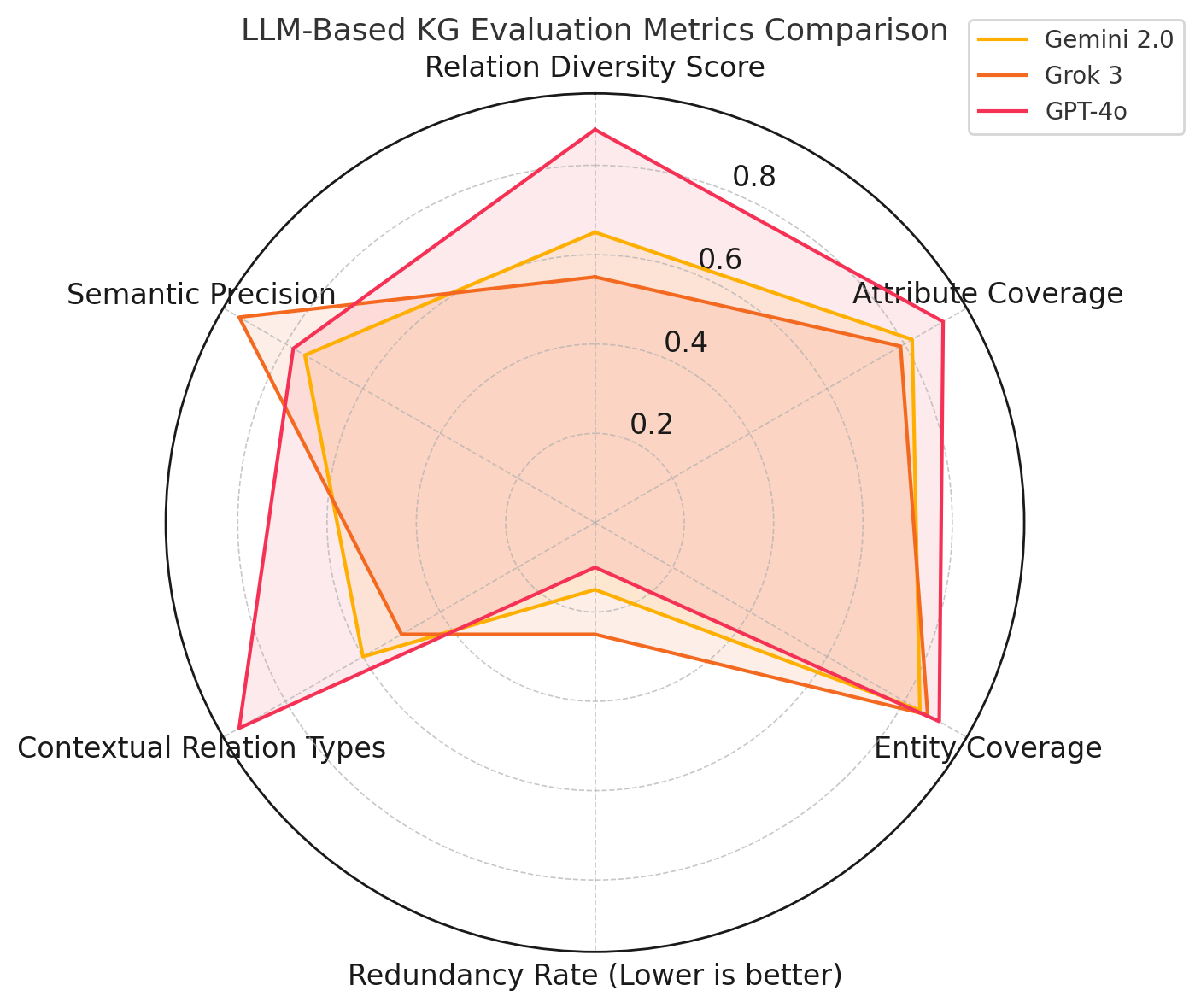
We compared relation types, source coverage, and structural patterns in knowledge graphs generated by Gemini 2.0 Flash, Grok 3, and GPT-4o across 40 oncology reports. Figure 4(a) summarizes key evaluation metrics.
Gemini and Grok focused on core clinical predicates (e.g., indicates,
treats, influences), while GPT-4o generated a broader range of
descriptive and context-sensitive relations (e.g., pertains_to,
assesses, documents), reflecting its strength in semantic
generalization.
All models extracted key clinical entities (Patient, Condition,
CarePlan) and biomarkers (age, ca_19_9, tumor_grade). GPT-4o
also surfaced nuanced attributes (e.g., fertility_preservation,
estrogen_exposure), showing sensitivity to subtle contextual cues
often missed by the others.
Structurally, each model connected densely around nodes like Condition
and Treatment. Gemini emphasized procedural clusters (e.g.,
Assessment, Practitioner); GPT-4o incorporated diverse concepts
(e.g., Therapy, FollowUp) and minimized redundancy. Grok prioritized
precision but exhibited more repetition due to conservative extraction.
Figure 4(a) illustrates:
GPT-4o excels in relational breadth and abstraction; Grok 3 in semantic
precision and redundancy control; and Gemini 2.0 in balanced procedural
accuracy.
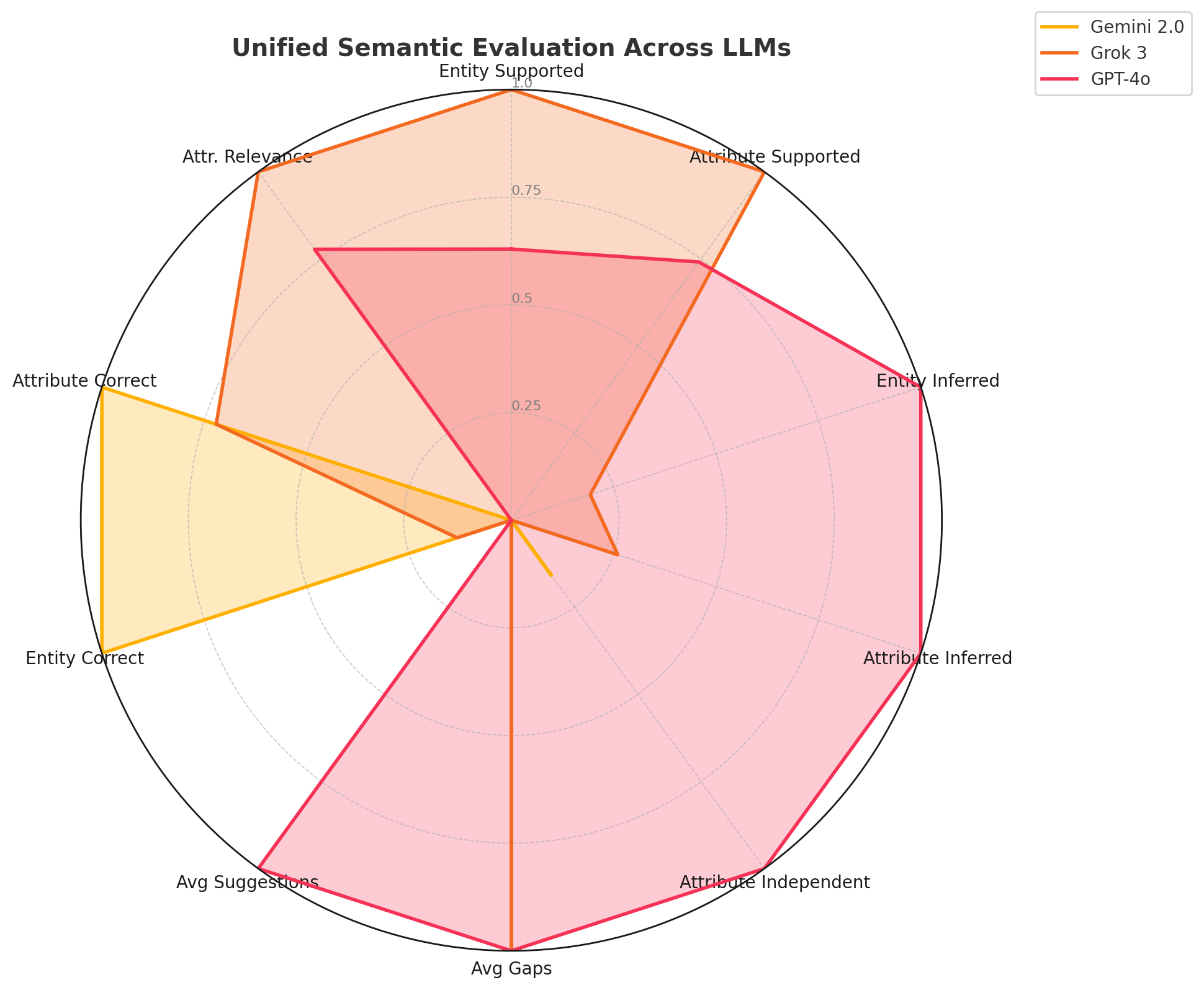
| Category | Metric | Gemini | Grok | GPT-4o | Example / Description |
|---|---|---|---|---|---|
| Data Support | Entity supported (per report) | 21.57 | 24.00 | 23.10 | Entities per patient
(Patient, Condition,
ImagingStudy). |
| Attribute supported (per report) | 67.67 | 89.03 | 83.50 | Examples: tumor_size,
Ki-67_Index, weight_loss. |
|
| Attribute diversity (Unique/Total) | 0.52 | 0.60 | 0.68 | Range of unique attributes, e.g., both
age and lymph_node_count. |
|
| Inference/Abstraction | Entity inferred (avg) | 0.12 | 0.28 | 0.95 | Infers HER2 implies a
Biomarker entity. |
| Attribute inferred (avg) | 0.70 | 1.35 | 3.20 | Infers Prognostic_Marker from
context. |
|
| Attribute independent (no entity) | 0.25 | 0.10 | 1.05 | Obesity extracted without
linked Patient. |
|
| Implicit inference ratio (%) | 1.01% | 1.49% | 3.92% | Implicit triple: ER-positive
→ responds_to → Tamoxifen. |
|
| Cross-sentence inference (#) | 1.0 | 1.5 | 3.2 | Combines scattered mentions of
surgery and blood_loss. |
|
| Gaps/Suggestions | Avg gaps per patient | 1.00 | 2.00 | 2.00 | Missing link: Diagnosis → associated_with → Biopsy. |
| Avg suggestions per patient | 1.00 | 1.00 | 1.50 | Suggests liver_function for
elevated bilirubin. |
|
| Gap-fill accuracy (%) | 55.0% | 61.0% | 68.0% | Fraction of suggestions matching expert-annotated content. | |
| Correctness/Relevance | Entity correct (%) | 99.4% | 23.3% | 11.7% | Correctly identifies
ImagingStudy as a clinical entity. |
| Attribute correct (%) | 97.5% | 75.3% | 18.0% | Example: CA_19-9 used
properly as LabResult. |
|
| Attribute relevance (avg) | 0.80 | 0.89 | 0.87 | Clinical utility of BMI and
Tumor_Grade. |
|
| Attribute inferred (%) | 0.95% | 0.18% | 2.49% | Infers Smoking_Status affects
Cancer_Risk. |
|
| Attribute independent (%) | 0.34% | 0.00% | 1.21% | Isolated attributes not linked to any entity. | |
| Triple validation confidence | 0.96 | 0.91 | 0.88 | Confidence in Biopsy → confirms → TumorType. |
|
| Semantic Quality | Hallucination rate (%) | 0.18% | 0.04% | 0.92% | False triple: Vitamin_D → prevents → Cancer. |
| Redundancy rate (%) | 2.2% | 0.7% | 1.8% | Relation repetition for the same attribute
(e.g., indicates). |
|
| SPARQL-compatible triples (%) | 98.1% | 99.6% | 96.2% | Fraction of triples executable in SPARQL endpoints. |
Semantic Relation Analysis Across LLMs
To evaluate the semantic integrity of relationships in the clinical
knowledge graph, we conducted a comparative analysis of predicate usage
by Gemini 2.0 Flash, Grok 3, and GPT-4o. Rather than focusing
solely on factual correctness, this analysis emphasizes whether each
model employs predicates that are both clinically meaningful and
ontologically valid (e.g., treats, leads_to, indicates). We
identified and grouped common relational errors into four categories:
(1) Vagueness, involving imprecise predicates such as
drug_use vague; (2) Causal Overreach, where unsupported causality is
overstated (e.g., bp leads_to comorbidities); (3) Incorrect
Semantics, such as the misuse of contraindicates or is_a; and (4)
Overuse/Generality, with excessive use of broad predicates like
indicates and influences.
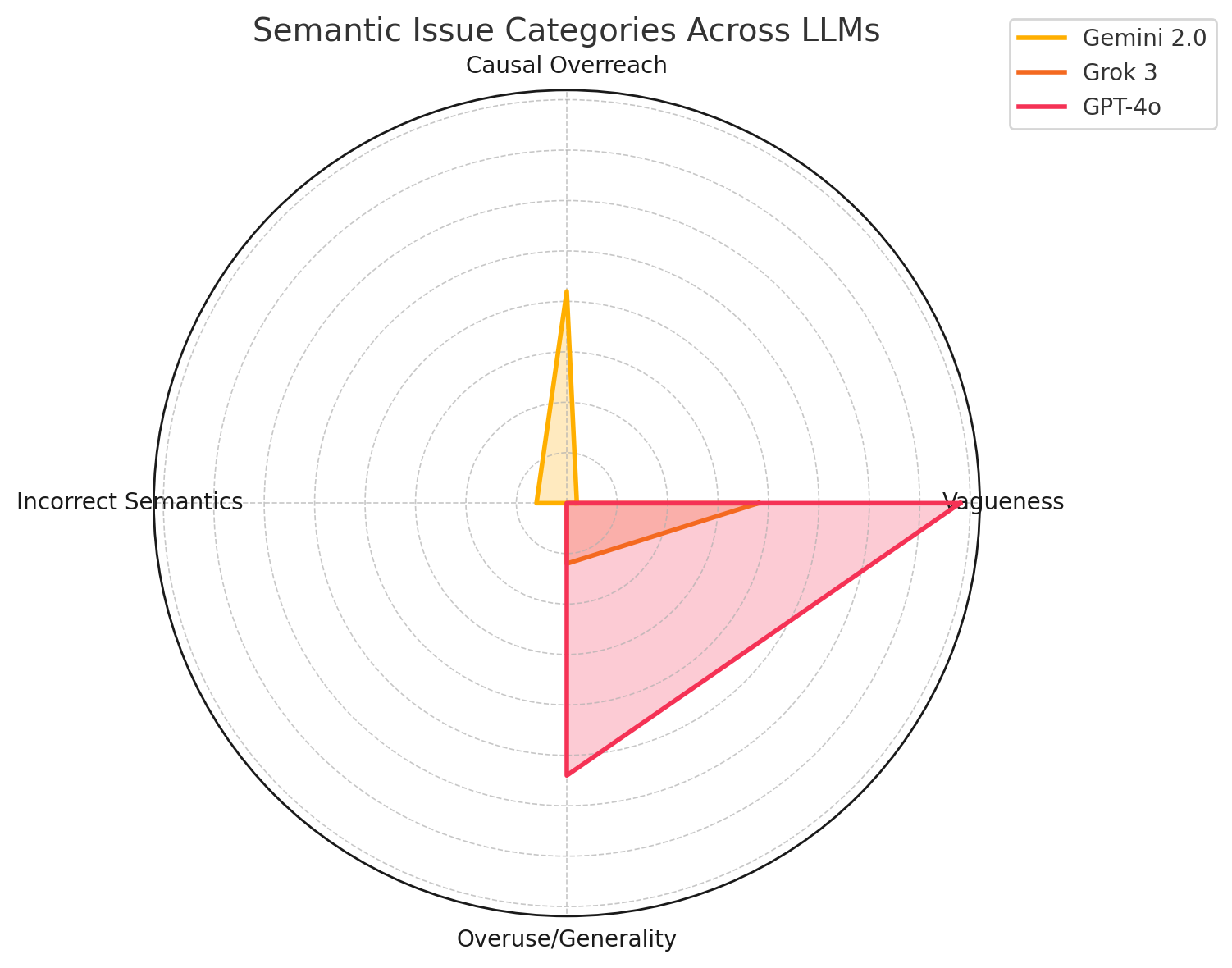
Figure 4(b) presents a radar
chart summarizing semantic issues. GPT-4o shows more vagueness and
generality due to its generative style; Grok 3 minimizes semantic errors
through rigorous filtering; Gemini 2.0 exhibits more causal overreach
from broader predicate exploration. These trends suggest that GPT-4o
provides relational diversity but may lack precision, Grok 3 emphasizes
semantic clarity and restraint, and Gemini 2.0 contributes exploratory
richness with some risk of drift. Together, their combined strengths
enable predicate refinement and clinical coherence in graph
construction.
Clinical Relevance Evaluation Across LLMs
To assess the clinical relevance of LLM-generated relationships, we compared average entity and attribute relevance across 40 oncology reports. As shown in Table [tab:llm_unified_summary_examples], GPT-4o achieved the highest relevance (0.87 for entities and 0.89 for attributes), followed by Grok (0.85 entity, 0.88 attribute), and Gemini (0.80 entity, 0.79 attribute). GPT-4o’s strength lies in contextual reasoning; Grok excels in conservative filtering; Gemini trades precision for broader coverage. These complementary capabilities make the LLM trio well-suited for building clinically grounded knowledge graphs that balance accuracy and discovery, and can be iteratively improved through expert feedback and retrieval-based refinement.
Unified Evaluation of LLM Behavior in KG Construction
We evaluated Gemini 2.0 Flash, Grok 3, and GPT-4o across six dimensions of clinical knowledge graph construction: data support, inference, gap handling, correctness, semantic quality, and SPARQL compatibility. Table [tab:llm_unified_summary_examples] provides a detailed comparison with clinical examples. Gemini 2.0 excels in precision, achieving the highest entity (99.4%) and attribute correctness (97.5%), and high SPARQL compliance (98.1%), though with limited inference and attribute diversity due to its schema-constrained approach.
Grok 3 offers strong semantic rigor and moderate inference, with high
attribute relevance (0.89), minimal hallucination (0.04%), and low
redundancy—ideal for conservative, high-precision graph construction.
GPT-4o leads in contextual inference and semantic enrichment,
extracting diverse attributes (e.g., estrogen_exposure) and achieving
the highest gap fill accuracy (68.0%). While hallucination is slightly
higher, its strength lies in uncovering implicit relations and improving
graph expressiveness. In the absence of ground truth, our multi-agent
ensemble ensures robustness by combining Gemini’s structured extraction,
Grok’s semantic validation, and GPT-4o’s contextual enrichment. This
yields ontology-aligned, clinically meaningful graphs suitable for
decision support and advanced analytics.
Limitations and Conclusion
Despite its strengths, the framework has several limitations. It is not yet integrated with live clinical decision support systems (CDSS), limiting real-world deployment. It currently operates solely on unstructured clinical text, omitting other critical modalities such as imaging (CT, MRI), biosignals (ECG, EMG), and structured EHR data. Our evaluation, based on 40 oncology reports (PDAC and BRCA) from the CORAL dataset, also limits generalizability across broader clinical settings. While model consensus and ontology alignment provide a form of weak supervision, expert-in-the-loop validation remains essential for resolving ambiguous or domain-specific triples.
We introduce the first framework to construct and evaluate clinical knowledge graphs directly from free-text reports using multi-LLM consensus and a Retrieval-Augmented Generation (RAG) strategy. Our pipeline combines schema-guided EAV extraction via Gemini 2.0 Flash, semantic refinement by GPT-4o and Grok 3, and ontology-aligned encoding using SNOMED CT, RxNorm, LOINC, GO, and ICD in RDF/RDFS/OWL with SWRL rules.
This KG-RAG approach supports continuous refinement, enabling iterative improvement of graph quality through self-supervised validation and expert feedback. By retrieving, validating, and grounding extracted triples over time, the system evolves dynamically, bridging static KG generation with adaptive knowledge refinement. Gemini anchors high-precision extraction, Grok filters hallucinations and enforces semantic rigor, and GPT-4o contributes contextual depth and abstraction, yielding clinically relevant, SPARQL-compatible graphs.
Future directions include real-time CDSS integration, multimodal fusion, large-scale validation across institutions, and enriched temporal and causal modeling. Ultimately, our framework provides a scalable and explainable foundation for transforming unstructured narratives into trustworthy, ontology-grounded clinical knowledge graphs.
📊 논문 시각자료 (Figures)





