Enhancing Histopathological Image Classification via Integrated HOG and Deep Features with Robust Noise Performance
📝 Original Paper Info
- Title: Enhancing Histopathological Image Classification via Integrated HOG and Deep Features with Robust Noise Performance- ArXiv ID: 2601.01056
- Date: 2026-01-03
- Authors: Ifeanyi Ezuma, Ugochukwu Ugwu
📝 Abstract
The era of digital pathology has advanced histopathological examinations, making automated image analysis essential in clinical practice. This study evaluates the classification performance of machine learning and deep learning models on the LC25000 dataset, which includes five classes of histopathological images. We used the fine-tuned InceptionResNet-v2 network both as a classifier and for feature extraction. Our results show that the fine-tuned InceptionResNet-v2 achieved a classification accuracy of 96.01\% and an average AUC of 96.8\%. Models trained on deep features from InceptionResNet-v2 outperformed those using only the pre-trained network, with the Neural Network model achieving an AUC of 99.99\% and accuracy of 99.84\%. Evaluating model robustness under varying SNR conditions revealed that models using deep features exhibited greater resilience, particularly GBM and KNN. The combination of HOG and deep features showed enhanced performance, however, less so in noisy environments.💡 Summary & Analysis
1. **Integration of HOG and Deep Features**: - This study introduces a novel method that combines HOG features with deep learning features for improved classification accuracy. - Metaphor: Think of this as combining the sharp vision of a hawk (HOG) to spot edges and textures, with the eagle's ability to see broader patterns from high above (deep learning), making the analysis more precise.-
Robustness and Resilience:
- The study evaluates the model’s robustness under varying conditions, particularly in terms of noise and signal-to-noise ratio.
- Metaphor: This method is like a bicycle that can handle rough terrain; it maintains high performance even with low SNR.
-
Optimization of Learning Rate and Loss Function:
- The study optimizes the learning rate dynamically to improve model performance during training.
- Metaphor: Adjusting the learning rate is similar to adjusting your speed while cycling – faster on a highway, slower in city traffic, ensuring you reach your destination safely.
📄 Full Paper Content (ArXiv Source)
Digital pathology has revolutionized histopathological examinations,
becoming a crucial technology in contemporary clinical practice and
increasingly essential within laboratory settings [1,2]. Modern
histopathological images are huge, containing up to billions of pixels
[3], and present complex features that are often too intricate to be
represented by simple mathematical equations or models [4]. This
complexity has driven the development of innovative automated methods
for image analysis, facilitating more accurate and efficient
diagnostics. Advances in Machine Learning (ML) and Artificial
Intelligence (AI) have further enhanced the automation of these
approaches, enabling the extraction of meaningful patterns from vast
amounts of image data. Presently, researchers and clinicians can now
leverage sophisticated algorithms and Deep Learning (DL) models to
analyze complex datasets with greater precision, facilitating early
disease detection and personalized treatment plans.
Traditional ML techniques for histopathological image analysis typically
involve several preprocessing steps, including feature selection, image
segmentation, and classification [5]. These methods have been
extensively reviewed in various literature as in [6][7][8].
However, over the last decade, there has been a notable shift towards
the development and deployment of DL techniques to enhance and optimize
complex biomedical image analysis. Specifically, after a successful
application of Convolutional Neural Networks (CNNs) in the 2012 ImageNet
Large Scale Visual Recognition Challenge [9 - 11], new open challenges
like CAMELYON16 [12][13] for lymph node metastasis detection and
PANDA [14] for Gleason grading in prostate cancer were introduced.
These challenges resulted in significant advancements, demonstrating
that classification systems based on CNN and Multiple-Instance Learning
(MIL) could outperform pathologists in these diagnostic tasks [15].
This shift underscores the increasing relevance of DL techniques in
biomedical imaging, highlighting their ability to deliver superior
performance in complex diagnostic scenarios.
Related works
In recent years, pathologists have benefited from extensive new
resources that have enhanced their approach to patient healthcare,
consequently elevating the associated outcomes [16]. Building on this
progress, researchers have carried out investigations into segmenting
different structures in breast histopathological images, employing
methods such as thresholding, fuzzy c-means clustering, and adaptive
thresholding, attaining results that vary in their level of success
[17 - 20]. Further advancements in the field include the use of
Support Vector Machine (SVM) to classify skin biopsies as melanoma or
nevi based on hematoxylin & eosin (H&E) staining. An SVC model was
developed using four features and, when tested under various
magnifications, performed best at 40x with the 3+4 feature combination,
achieving 90% accuracy [21]. In [22], the authors implemented
various feature extraction techniques, including VGG16, InceptionV3, and
ResNet50, to create eight distinct pre-trained CNN models for lung and
colon cancer classification. These models achieved accuracy rates
between 96% and 100%.
Additionally, the study in [23] utilized Otsu thresholding,
morphological operations, and histological constraints for segmenting
Whole Slide Images (WSIs). Machine Learning models, including Random
Forest (RF), K-Nearest Neighbors (KNN), Support Vector Machine (SVM),
and logistic regression, were trained with features extracted from
segmented image patches. The Random Forest model achieved the highest
accuracy at 93%. Moreover, Yan et al. [24] developed a novel hybrid
deep neural network combining convolutional and recurrent structures for
classifying breast cancer histopathological images into four categories.
Their method, which divided images into 12 patches for feature
extraction using Inception-V3, achieved 91.3% accuracy, outperforming
current methods. The study detailed in [25] focused on classifying
histopathology images of lung cancer using the LC25000 dataset and
convolutional neural networks (CNNs). Feature extraction was conducted
with ResNet50, VGG19, InceptionResNetV2, and DenseNet121. To improve
performance, the Triplet loss function was applied. The classification
task utilized a CNN with three hidden layers. Among the models,
Inception-ResNetv2 achieved the highest test accuracy rate, reaching
99.7%.
Contributions
This paper makes significant contributions through the following
innovative methodologies:
i. Integration of HOG and Deep Features for Enhanced Classification:
The primary contribution of this study is the introduction of a novel
method that integrates Histogram of Oriented Gradients (HOG) features
with deep features extracted from the InceptionResNet-v2 network. This
combined feature set leverages the strengths of both traditional HOG
features, which capture critical edge and texture information, and deep
learning features, which provide high-level abstract representations of
the images. This approach significantly improves classification accuracy
compared to conventional methods. This method stands out in the
literature as it offers a robust alternative to traditional methods,
such as LBP and Haralick, which have shown lower performance in previous
studies [26].
ii. Demonstrated Robustness and Resilience: The study also evaluates
the robustness of the proposed models under varying conditions,
particularly in terms of noise and signal-to-noise ratio (SNR). The
models trained on deep features exhibited greater resilience to these
variations, ensuring high performance even in less-than-ideal imaging
conditions. The findings suggest that the proposed integrated method not
only enhances accuracy but also maintains high reliability, which is
contrary to existing approaches in this field [27].
Material and Methods
Each experiment involved utilizing the provided architecture of InceptionResNet-V2. By leveraging this pre-trained network, we evaluated its performance in two distinct scenarios: first, as a standalone classifier and second, as a feature extractor. In the classification task, we directly applied InceptionResNet-V2 to categorize the images. For feature extraction, we used the network to obtain high-level features from the images, which were then fed into additional machine learning models for the final classification.
Dataset
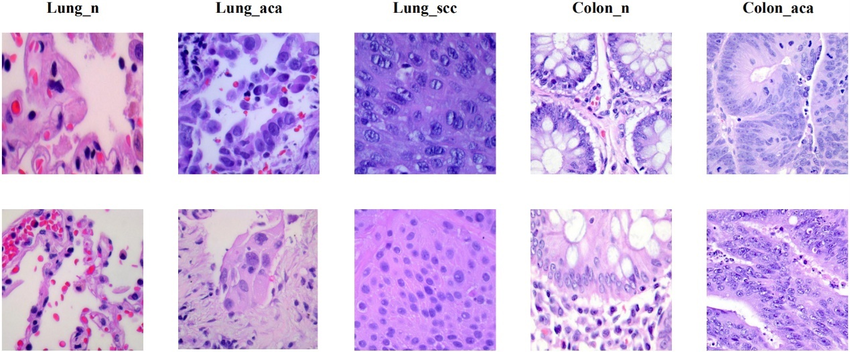
The LC25000 dataset contains 25,000 de-identified, HIPAA-compliant, and validated color images [28]. It comprises five classes: colon adenocarcinoma, benign colonic tissue, lung adenocarcinoma, lung squamous cell carcinoma, and benign lung tissue, each containing 5,000 images. The images were initially cropped to 768 x 768 pixels before enhancements such as left and right rotations (up to 25 degrees, with a 1.0 probability) and horizontal and vertical flips (with a 0.5 probability) were applied [29]. The data was then split into 60% for training, 20% for validation, and 20% for testing. Figure 1 shows samples from each class in the dataset.
Fine-Tuned Pre-trained CNN as Classifier
During the fine-tuning process, two convolutional blocks were defined, each consisting of a convolutional layer, a batch normalization layer, a ReLU activation function, and a max pooling layer. The first convolutional layer is composed of 128 kernels of size 7x7, with padding set to 3 to preserve the spatial dimensions of the input image after the convolution operation. Subsequently, the second convolutional layer employs 256 filters of size 5x5, with the stride set to 2, which effectively reduces the feature map dimensions by a factor of 2. Following these layers, it’s a fully connected layer to integrate the learned features for the final classification task. For optimization, we implemented Stochastic Gradient Descent with a Momentum value of 0.9. The initial learning rate was set at $`10^{-4}`$, and a piecewise learning rate schedule was adopted to determine how the learning rate changes over epochs.
Pre-Trained CNN for Feature Extraction
The images were augmented through random rotations (0°, 90°, 180°, 270°) and translations ([-3, 3] pixels) to increase the diversity of the training set. Features were extracted from the augmented images using the ’avg_pool’ layer of the InceptionResNet-v2 network. Several ML models were then trained on these features using Bayesian optimization for hyperparameter tuning.
Integration of Hog Features with Features Extracted from Pre-Trained CNN
The images were initially cropped to a size of 299 x 299 pixels and augmented through random rotations and translations. HOG features were extracted using a cell size of 128 x 128 pixels, and deep features were obtained from the ’avg_pool’ layer of the InceptionResNet-v2 network. These features were then concatenated to form a comprehensive feature vector. The same ML models as in section 4.2 were then trained on these combined features, with hyperparameters optimized through Bayesian optimization.
Results
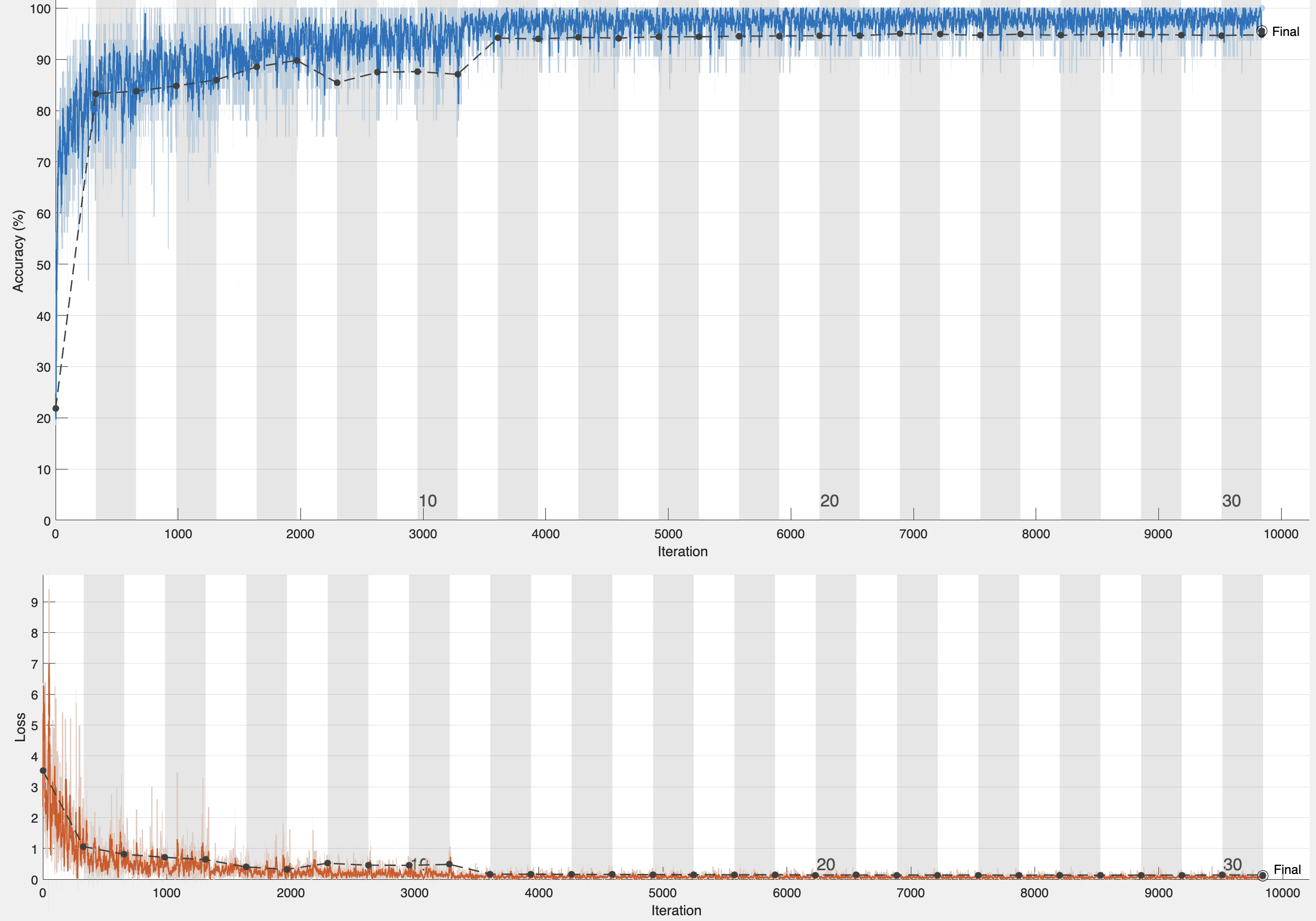
The learning rate and training loss, depicted in Figure 2(a), demonstrate the model’s rapid adaptation to the specific features of the histopathological images, with the training loss stabilizing as the optimal learning rate is achieved. Minor fluctuations in the training loss indicates
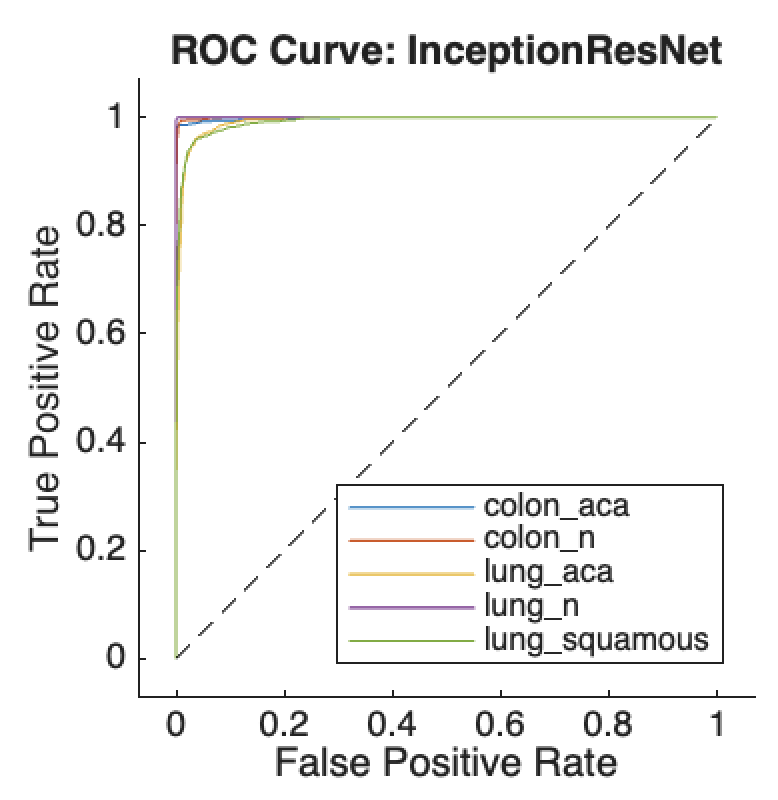
ongoing adjustments to the dataset’s intricate features, reflecting a dynamic learning process aimed at minimizing classification errors. The ROC curves for all five classes of histopathological images, shown in Figure 2(b), exhibit high true positive rates and low false positive rates across all classes. The model achieved a notable classification accuracy of 96.01%, with an average AUC of 96.8%, indicating strong overall performance.
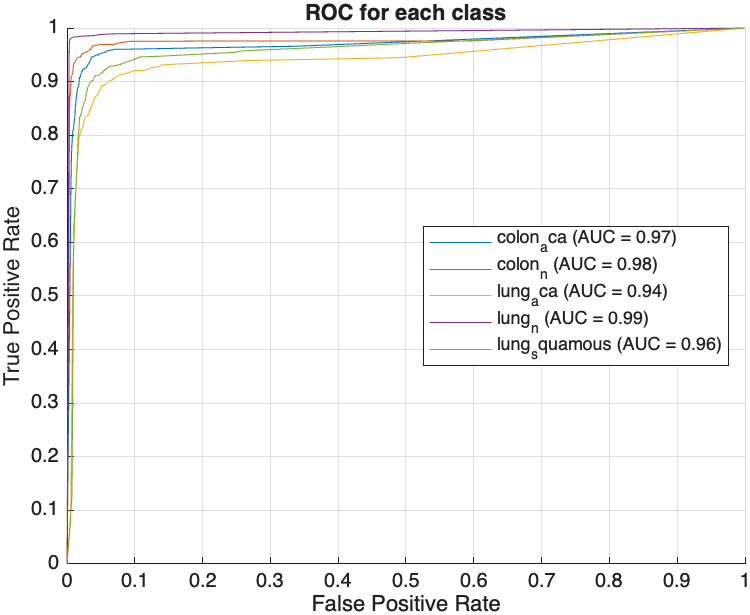
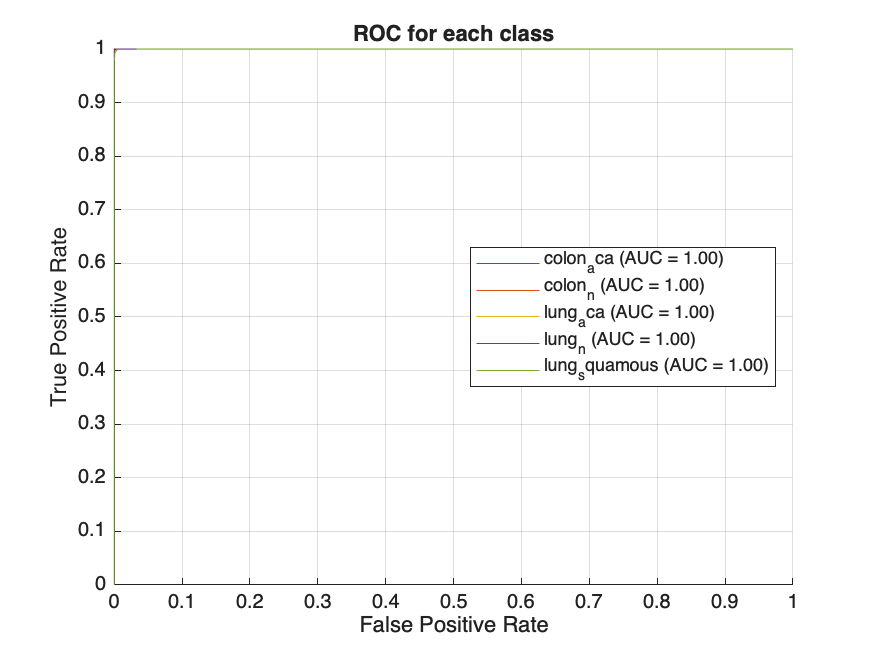
Following feature extraction, several machine learning models were trained on the augmented dataset, with hyperparameter tuning conducted via Bayesian optimization. The models evaluated included Decision Tree, Gradient Boosting Machine (GBM), k-Nearest Neighbors (KNN), Neural Network, and Support Vector Machine (SVM). The performance of these models was assessed based on two key metrics: AUC and test accuracy. The ROC curves obtained by the models are shown in Figure 3, with plots arranged from left to right, representing the worst to the best performing model. The Decision Tree model, illustrated in Figure 3(a), achieved an AUC of 95.85% and a test accuracy of 91.36%. While this indicates a solid performance, it is relatively lower compared to the other models evaluated. Significant improvements were observed with the GBM model, which achieved an AUC of 99.77% and a test accuracy of 98.05%. KNN, SVM, and Neural Network models demonstrated near-perfect performance, each achieving an AUC and test accuracies of over 99% as shown in Table 1. The Neural Network model, depicted in Figure 3(b), represents the best performing model in this experiment with an AUC of 99.99% and accuracy of 99.72%.
| Model | AUC(%) | Accuracy(%) |
|---|---|---|
| Decision Tree | 95.85 | 91.36 |
| GBM | 99.77 | 98.05 |
| KNN | 99.72 | 99.67 |
| Neural network | 99.99 | 99.72 |
| SVM | 99.94 | 99.03 |
Performance Metrics of Machine Learning Models on deep features extracted from InceptionResNet-V2.
The study also evaluated the performance of the different machine learning models using HOG features combined with deep features extracted from the pre-trained InceptionResNet-v2 network. When compared to the models trained using only deep features from InceptionResNet-v2, there was a noticeable improvement in performance across most models with the addition of HOG features as can be seen from Table 2. Among the models, the Decision Tree exhibited a
| Model | AUC(%) | Accuracy(%) |
|---|---|---|
| Decision Tree | 95.04 | 93.83 |
| GBM | 99.9 | 98.73 |
| KNN | 99.82 | 99.8 |
| Neural network | 99.99 | 99.84 |
| SVM | 99.96 | 99.40 |
Performance Metrics of Machine Learning Models on HOG and deep features extracted from InceptionResNet-V2.
solid but relatively modest performance, achieving an AUC of 95.04% and a test accuracy of 93.83%. In contrast, the GBM model demonstrated significant improvement with an AUC of 99.9% and a test accuracy of 98.73%. KNN and SVM models performed exceptionally well, with KNN achieving an AUC of 99.82% and an accuracy of 99.8%, while SVM attained an AUC of 99.96% and an accuracy of 99.40%. The Neural Network model emerged as the top performer, reaching an almost perfect AUC of 99.99% and a test accuracy of 99.84%. This suggests that combining HOG features with deep learning features enhances better classification performance.
Evaluating the Impact of Signal-to-Noise Ratio (SNR) on Model Performance
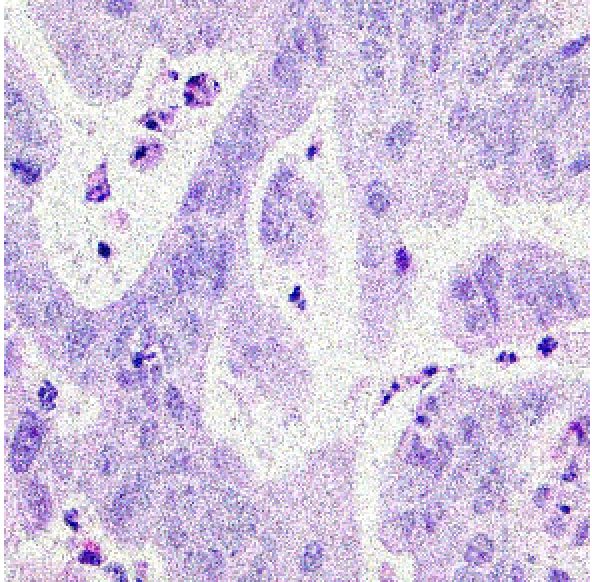
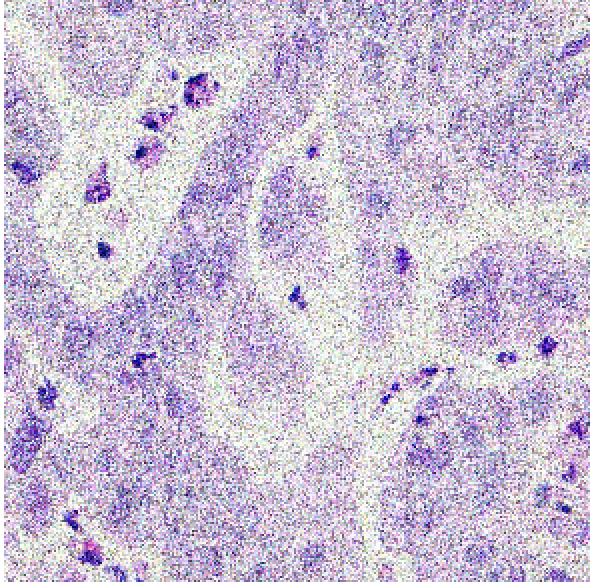
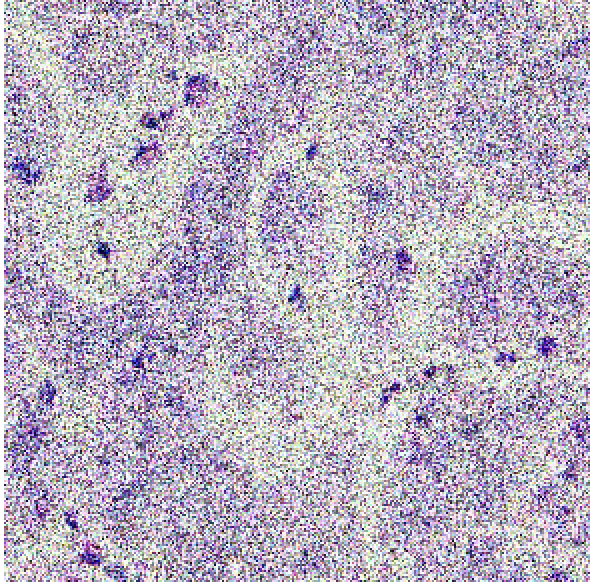
This section presents an in-depth evaluation of the impact of Signal-to-Noise Ratio (SNR) on the performance of all models that have been introduced in this study. To systematically assess the impact of SNR, we introduced different levels of noise to the dataset by applying additive Gaussian noise to the images at various SNR levels to simulate different noise conditions. The SNR levels were set at 40dB, 35dB, and 30dB to represent low, moderate, and high noise environments, respectively as shown in figure 4 below. These noisy datasets were then used to evaluate the robustness and adaptability of the models trained using the different methods described in section 3.2, 3.3 and 3.4. The impact of SNR on model performance was systematically compared across the three methods to evaluate the robustness and reliability of each model.
| 40dB | 35dB | 30dB |
 |
 |
 |
For the InceptionResNet-V2 used directly as a classifier, performance significantly deteriorated as noise levels increased, with accuracy dropping from 80.16% at 30dB to 27.44% at 40dB, as shown in Table 1. In contrast, models using deep features extracted from InceptionResNet-V2 exhibited more resilient performance under varying noise conditions. As shown in Figure 2(a),
| Method | SNR(dB) | Accuracy(%) |
|---|---|---|
| InceptionResNet-V2 as a classifier | 30 | 27.44 |
| 35 | 65.71 | |
| 40 | 80.16 |
the Decision Tree model’s accuracy improved from approximately 50% at 30dB to about 75% at 40dB, while more sophisticated models like GBM, KNN, Neural Network, and SVM achieved accuracies above 85% at 40dB. However, at 30dB, GBM attained an accuracy above 80%, with KNN following closely at 75.04%, while the Neural Network and SVM achieved significantly lower accuracies of 71.65% and 66.53%, respectively. On the other hand, with the combination of HOG features with deep features, as illustrated in Figure 2(b), all models attained similar levels of accuracy at 40dB compared with the performance of the models using deep feature extraction alone. Furthermore, there was little improvement observed with the Decision Tree
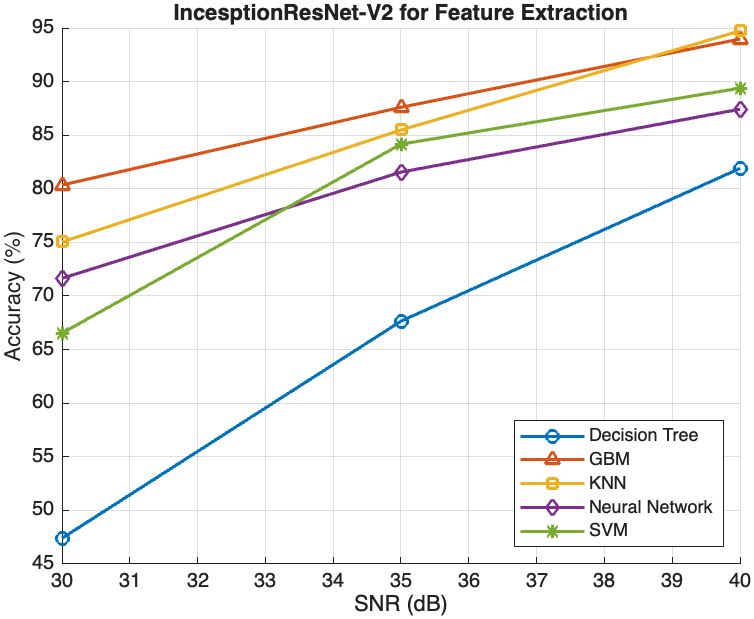
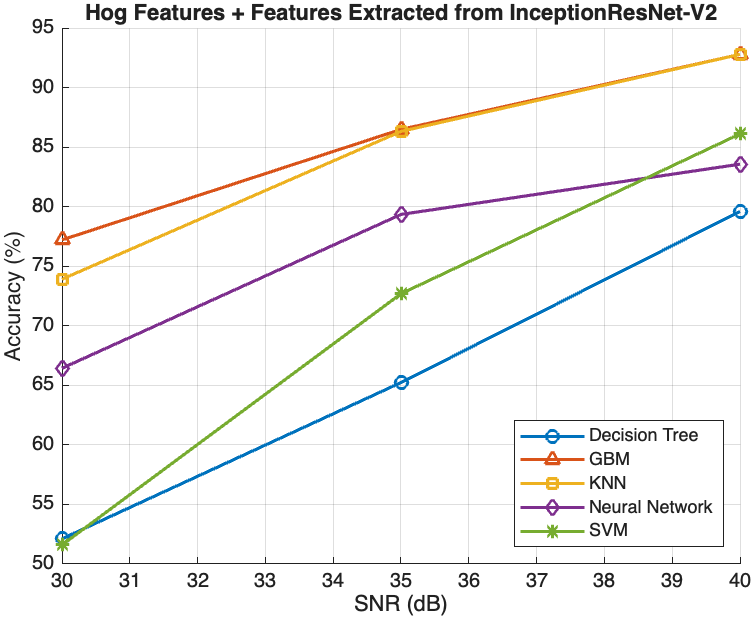
model, which achieved accuracies of 65.23% and 52.11% at 35dB and 30dB, respectively. However, the other models experienced relatively lower accuracies at 35dB and 30dB noise levels compared with the deep feature extraction method. GBM performed the best, achieving accuracies of 86.49% at 35dB and 77.21% at 30dB, followed closely by KNN with accuracies of 86.42% and 73.21%, respectively. Moreover, while the Neural Network and SVM models also improved with higher SNR levels, their performance gains were less pronounced compared to GBM and KNN. At 35dB and 30dB, the Neural Network model’s accuracy was 79.35% and 66.43%, respectively, whereas the SVM model’s accuracy was 72.69% and 51.59%, respectively.
Conclusion
This study evaluated the effectiveness of various ML and DL models in
classifying histopathological images, focusing on feature extraction
techniques and the impact of SNR on model performance. The fine-tuned
InceptionResNet-v2 network achieved notable classification accuracy
(96.01%) and AUC (96.8%). Models trained on deep features from
InceptionResNet-v2 outperformed those using only the pre-trained
network, with the Neural Network model achieving an almost perfect AUC
of 99.99% and accuracy of 99.84%. The addition of HOG features further
enhanced performance, particularly for advanced models like GBM, KNN,
Neural Network, and SVM.
Evaluating the impact of SNR revealed that models using deep features
were more resilient to noise, with GBM and KNN maintaining high accuracy
even at lower SNR levels. For instance, GBM achieved above 80% even at
30dB, while KNN maintained around 75% accuracy under the same
conditions. The combination of HOG and deep features showed similar
performance trends, though with slightly lower accuracy compared to
using deep features alone. For example, the Neural Network model
achieved 79.35% accuracy at 35dB and 66.43% at 30dB with the combined
features. The Neural Network model consistently emerged as the top
performer across different feature extraction methods and noise levels,
demonstrating the strength of combining fine-tuned neural networks with
robust feature extraction techniques. Other models like GBM and KNN also
showed strong performance, particularly in noisy conditions, making them
suitable alternatives depending on the specific diagnostic task
requirements. Overall, the integration of advanced feature extraction
methods and robust machine learning models significantly enhances the
classification performance of histopathological images, even under
challenging conditions.
H. R. Tizhoosh and L. Pantanowitz, “Artificial intelligence and digital pathology: challenges and opportunities,” Journal of pathology informatics, vol. 9, no. 1, p. 38, 2018.
N. Farahani, A. V. Parwani, and L. Pantanowitz, “Whole slide imaging in pathology: advantages, limitations, and emerging perspectives,” Pathol Lab Med Int, vol. 7, pp. 23-33, 4321.
A. B. Hamida, M. Devanne, J. Weber, C. Truntzer, V. Derangère, F. Ghiringhelli, and C. Wemmert, “Deep learning for colon cancer histopathological images analysis,” Computers in Biology and Medicine, vol. 136, p. 104730, 2021.
S. Banerji and S. Mitra, “Deep learning in histopathology: A review,” Wiley Interdisciplinary Reviews: Data Mining and Knowledge Discovery, vol. 12, no. 1, p. e1439, 2022.
S. M. Ayyad, M. Shehata, A. Shalaby, M. Abou El-Ghar, M. Ghazal, M. El-Melegy, and A. El-Baz, “Role of AI and histopathological images in detecting prostate cancer: a survey,” Sensors, vol. 21, no. 8, p. 2586, 2021.
J. Deng, W. Dong, R. Socher, L. J. Li, K. Li, and L. Fei-Fei, “2009 IEEE conference on computer vision and pattern recognition,” IEEE conference on computer vision and pattern recognition, pp. 248-255, 2009.
O. Russakovsky, J. Deng, H. Su, J. Krause, S. Satheesh, S. Ma, and L. Fei-Fei, “Imagenet large scale visual recognition challenge,” International journal of computer vision, vol. 115, pp. 211-252, 2015.
A. Krizhevsky, I. Sutskever, and G. E. Hinton, “ImageNet classification with deep convolutional neural networks,” Communications of the ACM, vol. 60, no. 6, pp. 84-90, 2017.
B. E. Bejnordi, M. Veta, P. J. Van Diest, B. Van Ginneken, N. Karssemeijer, G. Litjens, and CAMELYON16 Consortium, “Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer,” Jama, vol. 318, no. 22, pp. 2199-2210, 2017.
P. Bandi, O. Geessink, Q. Manson, M. Van Dijk, M. Balkenhol, M. Hermsen, and G. Litjens, “From detection of individual metastases to classification of lymph node status at the patient level: the camelyon17 challenge,” IEEE transactions on medical imaging, vol. 38, no. 2, pp. 550-560, 2018.
W. Bulten, K. Kartasalo, P. H. C. Chen, P. Ström, H. Pinckaers, K. Nagpal, and M. Eklund, “Artificial intelligence for diagnosis and Gleason grading of prostate cancer: the PANDA challenge,” Nature medicine, vol. 28, no. 1, pp. 154-163, 2022.
G. Campanella, M. G. Hanna, L. Geneslaw, A. Miraflor, V. Werneck Krauss Silva, K. J. Busam, and T. J. Fuchs, “Clinical-grade computational pathology using weakly supervised deep learning on whole slide images,” Nature medicine, vol. 25, no. 8, pp. 1301-1309, 2019.
C. H. Huang and E. M. Kalaw, “Automated classification for pathological prostate images using AdaBoost-based Ensemble Learning,” in 2016 IEEE Symposium Series on Computational Intelligence (SSCI), IEEE, pp. 1-4, 2016.
S. Rashid, L. Fazli, A. Boag, R. Siemens, P. Abolmaesumi, and S. E. Salcudean, “Separation of benign and malignant glands in prostatic adenocarcinoma,” in Medical Image Computing and Computer-Assisted Intervention–MICCAI 2013: 16th International Conference, Nagoya, Japan, September 22-26, 2013, Proceedings, Part III 16, Springer Berlin Heidelberg, pp. 461-468, 2013.
S. Naik, S. Doyle, S. Agner, A. Madabhushi, M. Feldman, and J. Tomaszewski, “Automated gland and nuclei segmentation for grading of prostate and breast cancer histopathology,” in 2008 5th IEEE International Symposium on Biomedical Imaging: From Nano to Macro, IEEE, pp. 284-287, 2008.
A. S. Sultan, M. A. Elgharib, T. Tavares, M. Jessri, and J. R. Basile, “The use of artificial intelligence, machine learning and deep learning in oncologic histopathology,” Journal of Oral Pathology & Medicine, vol. 49, no. 9, pp. 849-856, 2020.
M. N. Gurcan, L. E. Boucheron, A. Can, A. Madabhushi, N. M. Rajpoot, and B. Yener, “Histopathological image analysis: A review,” IEEE reviews in biomedical engineering, vol. 2, pp. 147-171, 2009.
S. Naik, S. Doyle, M. Feldman, J. Tomaszewski, and A. Madabhushi, “Gland segmentation and computerized gleason grading of prostate histology by integrating low-, high-level and domain specific information,” in MIAAB workshop, Piscataway, NJ, USA: Citeseer, pp. 1-8, 2007.
P. S. Karvelis, D. I. Fotiadis, I. Georgiou, and M. Syrrou, “A watershed based segmentation method for multispectral chromosome images classification,” in 2006 International Conference of the IEEE Engineering in Medicine and Biology Society, IEEE, pp. 3009-3012, 2006.
S. Petushi, F. U. Garcia, M. M. Haber, C. Katsinis, and A. Tozeren, “Large-scale computations on histology images reveal grade-differentiating parameters for breast cancer,” BMC medical imaging, vol. 6, no. 1, pp. 1-11, 2006.
Osborne, J. D., Gao, S., Chen, W. B., Andea, A., & Zhang, C. (2011, March). Machine classification of melanoma and nevi from skin lesions. In Proceedings of the 2011 ACM Symposium on Applied Computing (pp. 100-105).
Garg, S., & Garg, S. (2020, December). Prediction of lung and colon cancer through analysis of histopathological images by utilizing Pre-trained CNN models with visualization of class activation and saliency maps. In Proceedings of the 2020 3rd Artificial Intelligence and Cloud Computing Conference (pp. 38-45).
Valkonen, M., Kartasalo, K., Liimatainen, K., Nykter, M., Latonen, L., & Ruusuvuori, P. (2017). Metastasis detection from whole slide images using local features and random forests. Cytometry Part A, 91(6), 555-565.
Yan, R., Ren, F., Wang, Z., Wang, L., Zhang, T., Liu, Y., … & Zhang, F. (2020). Breast cancer histopathological image classification using a hybrid deep neural network. Methods, 173, 52-60.
Baranwal, N., Doravari, P., & Kachhoria, R. (2022). Classification of histopathology images of lung cancer using convolutional neural network (CNN). In Disruptive Developments in Biomedical Applications (pp. 75-89). CRC Press.
Doğan, R. S., & Yılmaz, B. (2024). Histopathology image classification: highlighting the gap between manual analysis and AI automation. Frontiers in Oncology, 13, 1325271.
Tripathi, S., & Singh, S. K. (2020). Ensembling handcrafted features with deep features: an analytical study for classification of routine colon cancer histopathological nuclei images. Multimedia Tools and Applications, 79(47), 34931-34954.
Borkowski, A. A., Bui, M. M., Thomas, L. B., Wilson, C. P., DeLand, L. A., & Mastorides, S. M. (2019). Lung and colon cancer histopathological image dataset (lc25000). arXiv preprint arXiv:1912.12142.
Hamed, E. A. R., Salem, M. A. M., Badr, N. L., & Tolba, M. F. (2023). An efficient combination of convolutional neural network and LightGBM algorithm for lung cancer histopathology classification. Diagnostics, 13(15), 2469.
Liu, Y., Wang, H., Song, K., Sun, M., Shao, Y., Xue, S., … & Zhang, T. (2022). CroReLU: cross-crossing space-based visual activation function for lung cancer pathology image recognition. Cancers, 14(21), 5181.
📊 논문 시각자료 (Figures)