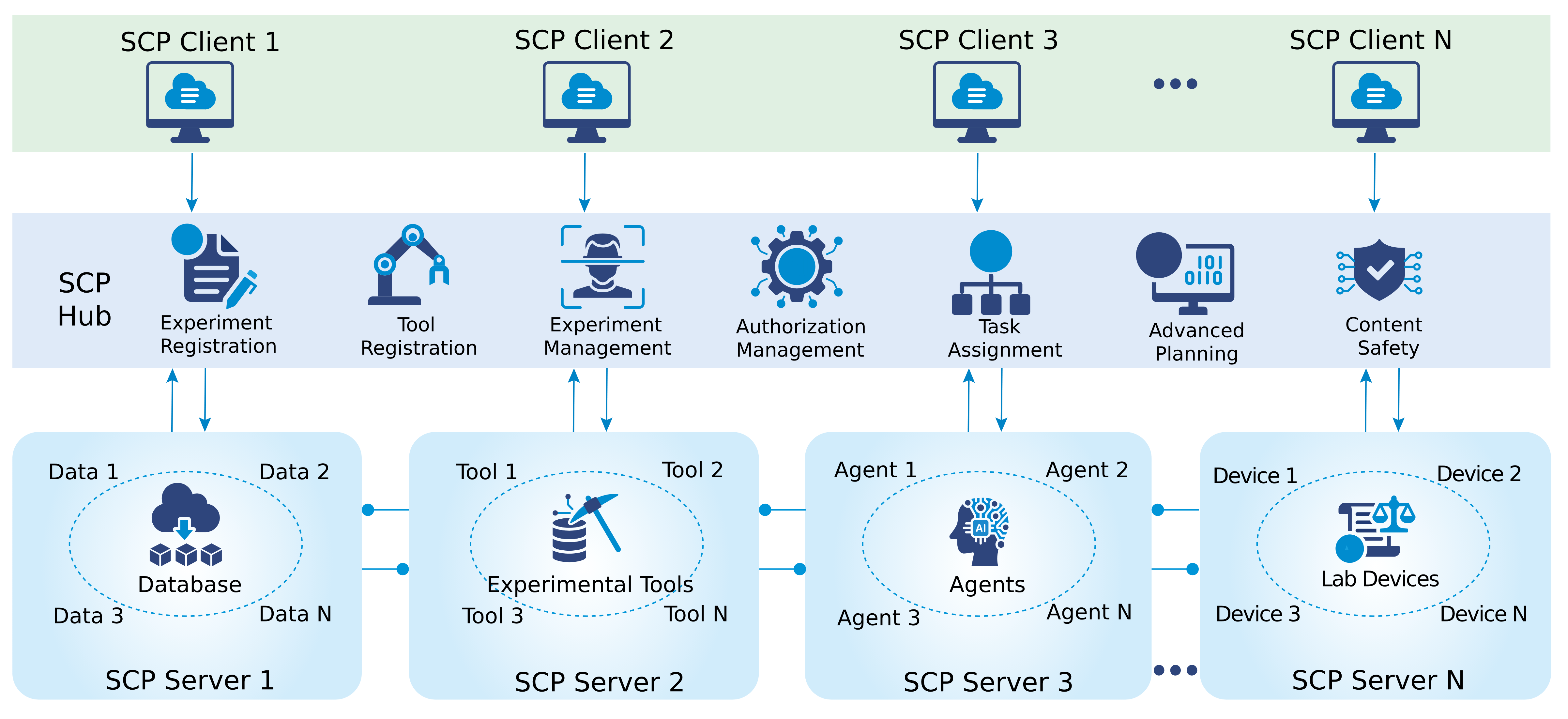
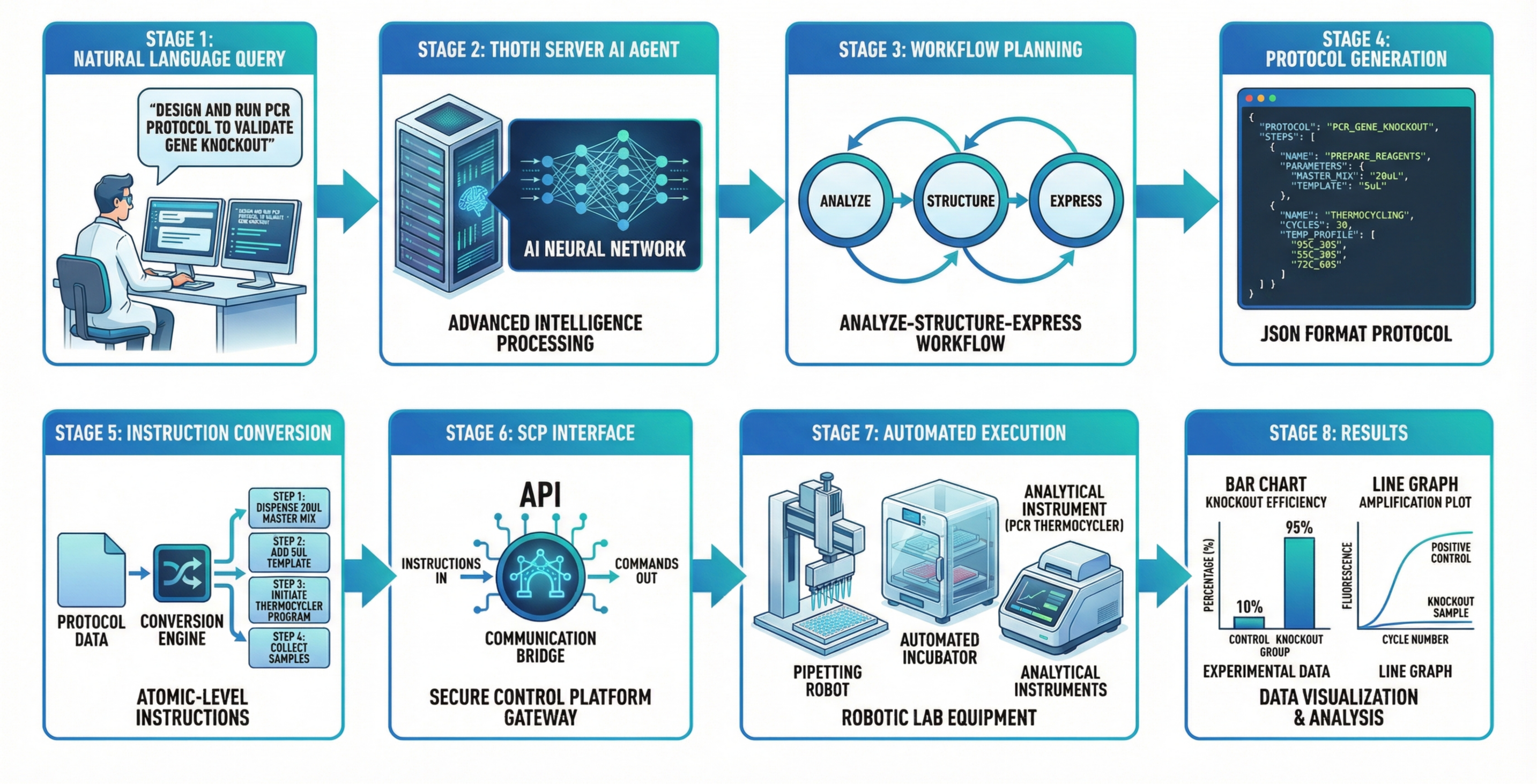
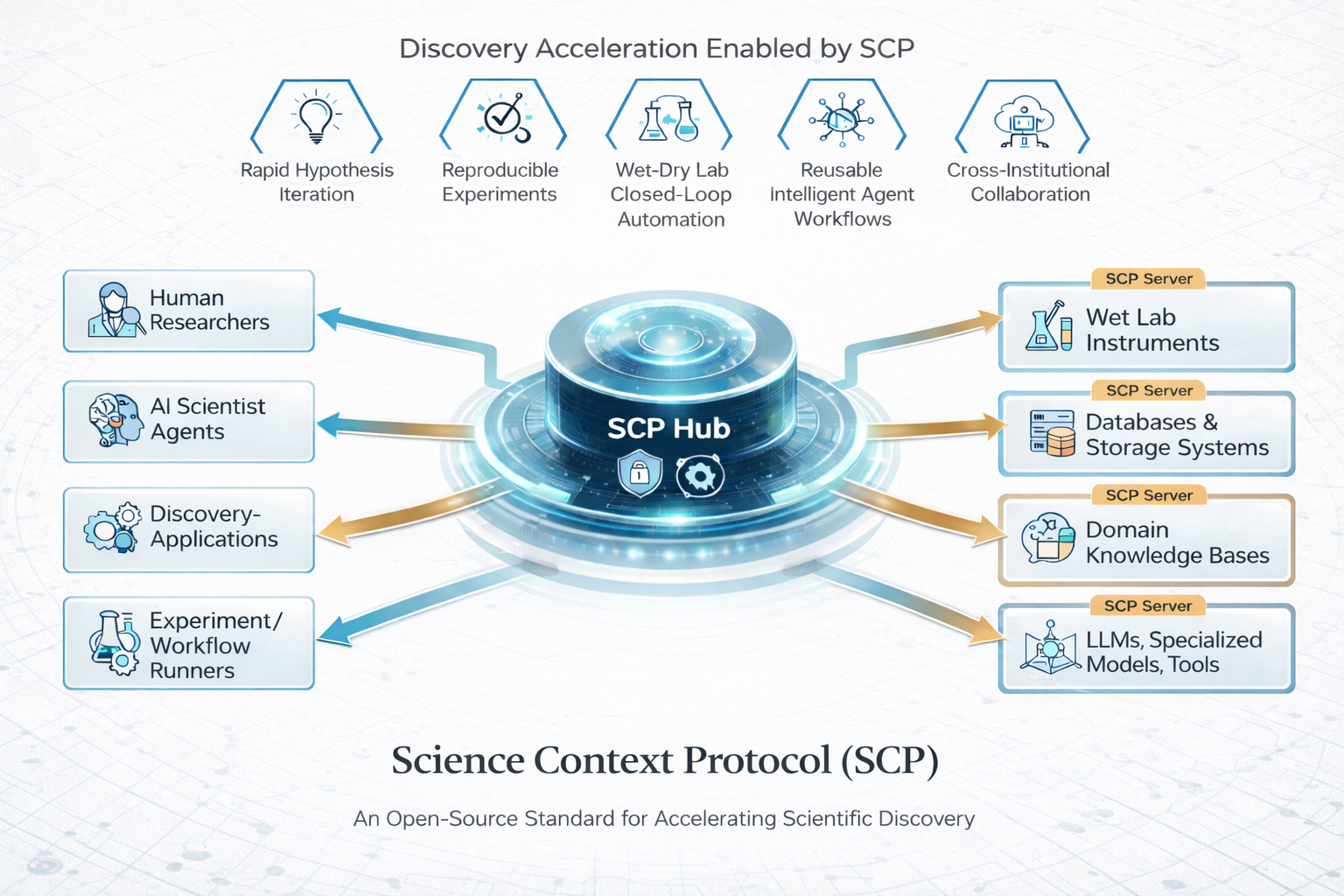
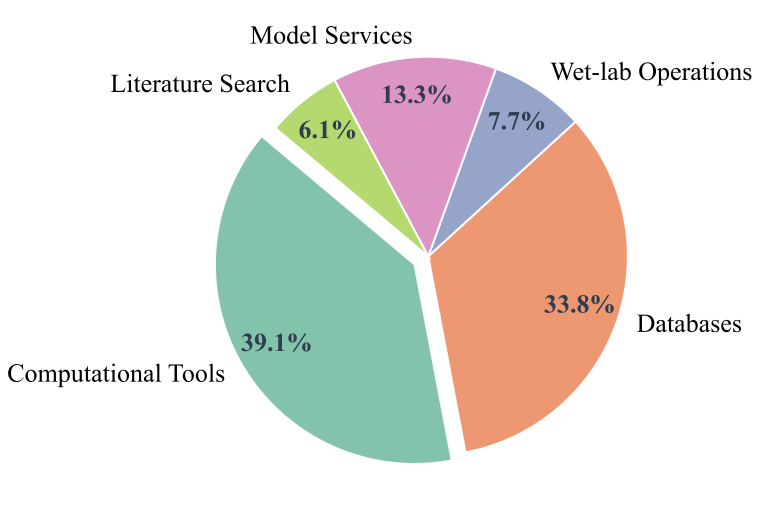
SCP Accelerating Discovery with a Global Web of Autonomous Scientific Agents
📝 Original Paper Info
- Title: SCP Accelerating Discovery with a Global Web of Autonomous Scientific Agents- ArXiv ID: 2512.24189
- Date: 2025-12-30
- Authors: Yankai Jiang, Wenjie Lou, Lilong Wang, Zhenyu Tang, Shiyang Feng, Jiaxuan Lu, Haoran Sun, Yaning Pan, Shuang Gu, Haoyang Su, Feng Liu, Wangxu Wei, Pan Tan, Dongzhan Zhou, Fenghua Ling, Cheng Tan, Bo Zhang, Xiaosong Wang, Lei Bai, Bowen Zhou
📝 Abstract
We introduce SCP: the Science Context Protocol, an open-source standard designed to accelerate discovery by enabling a global network of autonomous scientific agents. SCP is built on two foundational pillars: (1) Unified Resource Integration: At its core, SCP provides a universal specification for describing and invoking scientific resources, spanning software tools, models, datasets, and physical instruments. This protocol-level standardization enables AI agents and applications to discover, call, and compose capabilities seamlessly across disparate platforms and institutional boundaries. (2) Orchestrated Experiment Lifecycle Management: SCP complements the protocol with a secure service architecture, which comprises a centralized SCP Hub and federated SCP Servers. This architecture manages the complete experiment lifecycle (registration, planning, execution, monitoring, and archival), enforces fine-grained authentication and authorization, and orchestrates traceable, end-to-end workflows that bridge computational and physical laboratories. Based on SCP, we have constructed a scientific discovery platform that offers researchers and agents a large-scale ecosystem of more than 1,600 tool resources. Across diverse use cases, SCP facilitates secure, large-scale collaboration between heterogeneous AI systems and human researchers while significantly reducing integration overhead and enhancing reproducibility. By standardizing scientific context and tool orchestration at the protocol level, SCP establishes essential infrastructure for scalable, multi-institution, agent-driven science.💡 Summary & Analysis
This paper compiles a variety of tools extensively used in life sciences and engineering. It focuses particularly on protein function prediction, molecular structure visualization, and biological data processing. Each tool is specialized for specific computational or analytical tasks, significantly enhancing efficiency in solving complex problems within the fields of life sciences and engineering.📄 Full Paper Content (ArXiv Source)
| Tool Name | Description | Category |
| Tool Name | Description | Category |
| Predict protein function. | Computational Tools | |
| Predict functional residue. | Computational Tools | |
| Check if the input protein sequence string is valid. | Computational Tools | |
| Check if the input SMILES string is valid | Computational Tools | |
| Convert a list of SMILES strings or a list of SMI file paths into other molecular formats. | Computational Tools | |
| Convert a protein file from PDB format to PDBQT format for docking preparation. | Computational Tools | |
| Convert a protein-ligand complex file from CIF format to PDB format. | Computational Tools | |
| Visualize the protein structure and save as an image. | Computational Tools | |
| Visualize the molecular structure and save as an image. | Computational Tools | |
| Visualize the protein-ligand complex structure and save as an image. | Computational Tools | |
| Use PDBFixer to repair the protein structure PDB file in preparation for molecular docking. | Computational Tools | |
| Read the input smi file and extract the SMILES strings along with their corresponding compound names. | Computational Tools | |
| Parse protein sequences from the input fasta file. | Computational Tools | |
| Compute a set of basic molecular properties for each SMILES. | Computational Tools | |
| Compute hydrophobicity-related molecular descriptors for each SMILES. | Computational Tools | |
| Compute hydrogen bonding-related properties for each SMILES. | Computational Tools | |
| Compute a set of molecular complexity descriptors for each SMILES. | Computational Tools | |
| Compute a set of topological descriptors for each SMILES. | Computational Tools | |
| Compute key drug-likeness metrics for each SMILES. | Computational Tools | |
| Compute Gasteiger partial charges and formal charge for each SMILES. | Computational Tools | |
| Compute custom molecular complexity-related descriptors for each SMILES. | Computational Tools | |
| Compute a set of physicochemical properties for the input protein sequence. | Computational Tools | |
| Read a protein pdb file and compute basic structural statistics. | Computational Tools | |
| Read a protein pdb file and compute key geometric properties based on $`C_\alpha`$ atom coordinates. | Computational Tools | |
| Read a protein pdb file and compute three key quality indicators. | Computational Tools | |
| Read a protein pdb file and analyze compositional details by counting occurrences of each atom name, residue name, and the number of atoms per chain. | Computational Tools | |
| Compute the Tanimoto similarities between a target molecule and a list of candidate molecules using Morgan fingerprints. | Computational Tools | |
| Extract the specified chain or the longest amino-acid chain from the input protein structure file and save as a new PDB file. | Computational Tools | |
| Extract the amino acid sequence of each chain from the PDB file. | Computational Tools | |
| Convert files smaller than 10MB to Base64 encoding. | Computational Tools | |
| Convert Base64 encoding back to a file. | Computational Tools | |
| Use P2Rank to predict ligand binding pockets in the input protein. | Computational Tools | |
| Perform molecular docking using QuickVina2-GPU. | Computational Tools | |
| Enter a list of candidate small molecules. | Computational Tools | |
| Use Boltz to predict binding affinity between protein (receptor) and small molecules (ligands). | Computational Tools | |
| Use fpocket to predict protein pockets and set it as the default tool for pocket prediction. | Computational Tools | |
| Map CDS coordinates to genomic coordinates. | Computational Tools | |
| Translate between different variant nomenclature systems and representations. | Computational Tools | |
| Computes and returns LD values between the given variant and all other variants in a window. | Computational Tools | |
| Computes and returns LD values between the given variants. | Computational Tools | |
| Computes and returns LD values between all pairs of variants in the defined region. | Computational Tools | |
| Map coordinates between assemblies. | Computational Tools | |
| Computes observed transcript haplotype sequences based on phased genotype data. | Computational Tools | |
| Batch predict the functional effects of multiple variants using VEP with HGVS notation. | Computational Tools | |
| Get data from a specific track for a genomic region. | Computational Tools | |
| Comprehensive protein analysis combining InterProScan (domain analysis) and BLAST (similarity search). | Computational Tools | |
| Complete code to perform a certain calculation. | Computational Tools | |
| Perform a general validation/check operation. | Computational Tools | |
| Insert an item into a target. | Computational Tools | |
| Open an item. | Computational Tools | |
| Analyze real-time PCR run results using a specified analysis software. | Computational Tools | |
| Generate executable JSON file from protocol text. | Computational Tools | |
| Extract experimental protocol from a provided PDF Params: pdf_url (str): PDF file URL Returns: protocol (str): protocol text Arg: arguments (dict) - Tool parameter dictionary | Computational Tools | |
| Calculate the geometric term $`\sqrt{\pi a}`$. | Computational Tools | |
| Calculate the stress intensity factor KI. | Computational Tools | |
| Determine if fracture occurs. | Computational Tools | |
| Calculate the ratio of the new mobility relative to the original mobility. | Computational Tools | |
| Calculate the unit cell volume of a crystal with lattice constant a. | Computational Tools | |
| Calculate density as mass divided by volume. | Computational Tools | |
| Calculate the proportionality constant K from known diameter, length, and weight. | Computational Tools | |
| Calculate the moment of inertia of a ring. | Computational Tools | |
| Calculate the moment of inertia of a spoke (rod) about one end. | Computational Tools | |
| Calculate total moment of inertia of all spokes. | Computational Tools | |
| Calculate the weight of concrete. | Computational Tools | |
| Convert crack length to meters based on the specified unit. | Computational Tools | |
| Calculate critical stress using Griffith-Irwin equation. | Computational Tools | |
| Validate that the axial strain is non-zero. | Computational Tools | |
| Compute the material density based on specific gravity. | Computational Tools | |
| Calculate the volume in cubic meters given mass and density. | Computational Tools | |
| Calculate the angular frequency of a harmonic oscillator. | Computational Tools | |
| Calculate density in g/cm³. | Computational Tools | |
| Convert Young’s modulus from GPa to MPa. | Computational Tools | |
| Calculate the exponent used in the Mott VRH model based on system dimension. | Computational Tools | |
| Calculate the magnetic anisotropy energy. | Computational Tools | |
| Convert elastic modulus from GPa to Pa. | Computational Tools | |
| Calculate the stiffness contribution (E * A) for a material. | Computational Tools | |
| Sum individual stiffness contributions to get total stiffness. | Computational Tools | |
| Calculate tensile stress in Pascals. | Computational Tools | |
| Convert stress from Pascals to Megapascals. | Computational Tools | |
| Calculate the volume of the oil droplet. | Computational Tools | |
| Calculate atomic packing factor (APF). | Computational Tools | |
| Calculate percentage difference between two packing factors. | Computational Tools | |
| Validate that the stress unit is within the allowed units. | Computational Tools | |
| Calculate elastic modulus of alloy using rule of mixtures. | Computational Tools | |
| Calculate the moment of inertia of a hollow sphere about its diameter. | Computational Tools | |
| Calculate yield strength using Hall-Petch equation. | Computational Tools | |
| Calculate the strain (dimensionless) from elongation and original length. | Computational Tools | |
| Calculate the elastic energy density (J/m³) from Young’s modulus and strain. | Computational Tools | |
| Calculate the total elastic energy stored in the material. | Computational Tools | |
| Convert stress to Pascals (Pa). | Computational Tools | |
| Convert a stress value from Pascals to the specified output unit. | Computational Tools | |
| Compute the compression ratio. | Computational Tools | |
| Calculate mass from volume and density. | Computational Tools | |
| Calculate weight in Newtons. | Computational Tools | |
| Calculate buoyant force in Newtons. | Computational Tools | |
| Calculate total thickness in nanometers. | Computational Tools | |
| Calculate the transverse strain using Poisson’s ratio and longitudinal strain. | Computational Tools | |
| Calculate the length change due to thermal expansion. | Computational Tools | |
| Calculate the final length after thermal contraction. | Computational Tools | |
| Calculate the relative change in length as a percentage. | Computational Tools | |
| Calculate the minimum coating thickness needed for waterproofing ($`\theta \geq 90^{\circ}`$). | Computational Tools | |
| Calculate the atomic radius for BCC structure. | Computational Tools | |
| Calculate the density of a BCC structured material. | Computational Tools | |
| Calculate stress according to Hooke’s law. | Computational Tools | |
| Determine whether interface debonding will occur. | Computational Tools | |
| Calculate safety factor. | Computational Tools | |
| Convert stress from MPa to Pascals. | Computational Tools | |
| Calculate the plastic section modulus Z for a rectangular cross-section. | Computational Tools | |
| Calculate the plastic moment Mp in kN·m. | Computational Tools | |
| Compute the density from mass and volume. | Computational Tools | |
| Convert shear modulus from GPa to Pa. | Computational Tools | |
| Calculate the moment of inertia of a hollow cylinder about its central axis. | Computational Tools | |
| Calculate the final surface roughness after reduction. | Computational Tools | |
| Calculate the percentage increase in hardness. | Computational Tools | |
| Calculate impact toughness in J/$`m^2`$. | Computational Tools | |
| Convert impact toughness from J/$`m^2`$ to kJ/$`m^2`$. | Computational Tools | |
| Calculate volume occupied by suspended solids (m³/m²). | Computational Tools | |
| Calculate stress amplitude. | Computational Tools | |
| Calculate mean stress. | Computational Tools | |
| Calculate stress ratio R = $`\sigma_{min} / \sigma_{max}`$. | Computational Tools | |
| Compute the numerator for Jurin’s law: $`2\times \sigma \times cos(\theta)`$. | Computational Tools | |
| Sorts a list of three stresses in descending order. | Computational Tools | |
| Calculates the maximum shear stress from principal stresses. | Computational Tools | |
| Calculate the plastic zone radius. | Computational Tools | |
| Calculate specific surface area (SSA) based on constant k and diameter. | Computational Tools | |
| Calculate the interplanar spacing for a BCC crystal. | Computational Tools | |
| Check if the (h,k,l) plane is allowed for diffraction in BCC structure. | Computational Tools | |
| Calculate minimum grain size according to maximum corrosion rate and porosity. | Computational Tools | |
| Calculate the elongation of the specimen. | Computational Tools | |
| Determine if interface stress exceeds allowable stress. | Computational Tools | |
| Find the conductivity at zero strain. | Computational Tools | |
| Calculate the characteristic length as the ratio of diffusion coefficient to front velocity. | Computational Tools | |
| Convert Young’s modulus to Pascals. | Computational Tools | |
| Calculate the total mass of a material. | Computational Tools | |
| Calculate shear stress in pascals. | Computational Tools | |
| Assess safety based on shear stress and allowable stress. | Computational Tools | |
| Calculate the moment of inertia for a rectangular cross-section. | Computational Tools | |
| Return the theoretical packing factor for FCC. | Computational Tools | |
| Calculate the stress amplitude from maximum and minimum stresses. | Computational Tools | |
| Calculate the fatigue life (number of cycles) from stress ratio, fatigue strength coefficient, and exponent. | Computational Tools | |
| Determine if material is in elastic deformation stage. | Computational Tools | |
| Calculate effective elastic modulus of porous materials. | Computational Tools | |
| Assign default parameters for a given material type. | Computational Tools | |
| Calculate kd divided by sqrt(d). | Computational Tools | |
| Calculate the yield strength based on $`\sigma_0`$ and $`\frac{kd}{\sqrt{d}}`$. | Computational Tools | |
| Calculate the weight of the wire (gravitational force). | Computational Tools | |
| Calculate the moment of inertia for a solid sphere. | Computational Tools | |
| Determine which material has higher remaining strength. | Computational Tools | |
| Calculate the initial volume of a material. | Computational Tools | |
| Calculate the maximum compressive force. | Computational Tools | |
| Calculate the difference between fiber and matrix elastic moduli. | Computational Tools | |
| Calculate the contribution of fibers to the composite elastic modulus. | Computational Tools | |
| Calculate mass from density and volume. | Computational Tools | |
| Calculate the polar moment of inertia $`J = \frac{\pi}{2} \cdot r^4`$. | Computational Tools | |
| Calculate allowable shear stress, which is half of the yield strength. | Computational Tools | |
| Determine if plastic deformation will occur. | Computational Tools | |
| Calculate the volume of a material based on its mass and density. | Computational Tools | |
| Calculate surface roughness (Ra) based on thickness. | Computational Tools | |
| Calculate interface stress. | Computational Tools | |
| Convert resistance from kilo-ohms to ohms. | Computational Tools | |
| Calculate required voltage using Ohm’s Law V = I × R. | Computational Tools | |
| Calculate noise current. | Computational Tools | |
| Calculate the capacitance in Farads based on physical parameters. | Computational Tools | |
| Calculate the electric charge in a capacitor. | Computational Tools | |
| Calculate the capacitance after inserting dielectric. | Computational Tools | |
| Calculate voltage from charge and capacitance. | Computational Tools | |
| Calculate the cell potential (V) from cathode and anode potentials. | Computational Tools | |
| Compute the electrical conductivity using the Mott VRH model. | Computational Tools | |
| Calculate the electric field strength between two plates. | Computational Tools | |
| Calculate the total charge on the oil droplet. | Computational Tools | |
| Calculate the mass of the oil droplet based on electric force balance. | Computational Tools | |
| Calculate the charge stored on a capacitor. | Computational Tools | |
| Calculate the energy difference between two energy levels (in eV). | Computational Tools | |
| Calculate resistivity from conductivity. | Computational Tools | |
| Calculate the vacuum capacitance of a parallel plate capacitor. | Computational Tools | |
| Calculate the energy stored in a capacitor. | Computational Tools | |
| Calculate the electric potential at a given position ratio between plates. | Computational Tools | |
| Calculate the electric field strength produced by a point charge. | Computational Tools | |
| Calculate the force experienced by a charge in an electric field. | Computational Tools | |
| Calculate the equivalent resistance of resistors in parallel. | Computational Tools | |
| Calculate the maximum output voltage of a series-connected battery pack. | Computational Tools | |
| Calculate the duty cycle for a boost converter. | Computational Tools | |
| Calculate the magnitude of the induced EMF. | Computational Tools | |
| Calculate the voltage across a resistor using Ohm’s Law. | Computational Tools | |
| Calculate the current needed for a specified output power given the battery voltage. | Computational Tools | |
| Calculate total current consumption including quiescent and output currents. | Computational Tools | |
| Calculate the battery life in hours. | Computational Tools | |
| Calculate the RC time constant ($`\tau`$) for an RC circuit. | Computational Tools | |
| Calculate the absolute permittivity of a dielectric material. | Computational Tools | |
| Calculate the capacitance of a parallel plate capacitor. | Computational Tools | |
| Calculate the energy stored in a capacitor. | Computational Tools | |
| Calculate the current required to generate a specified heat energy in a resistor over a given time. | Computational Tools | |
| Calculate the vacuum magnetic permeability ($`\mu_o`$). | Computational Tools | |
| Calculate the magnetic flux density B. | Computational Tools | |
| Return the vacuum permittivity (epsilon_0) in F/m. | Computational Tools | |
| Convert energy to charge (Coulombs). | Computational Tools | |
| Convert Coulombs to milliamp-hours. | Computational Tools | |
| Calculate the RC time constant. | Computational Tools | |
| Calculate the collector current in a transistor circuit. | Computational Tools | |
| Calculate the emitter current in a transistor circuit. | Computational Tools | |
| Calculate voltage drop across a resistor. | Computational Tools | |
| Calculate the power supply voltage Vcc. | Computational Tools | |
| Calculate Vcc using the analytical formula for verification. | Computational Tools | |
| Calculate the total number of turns in a solenoid. | Computational Tools | |
| Calculate the number of turns per meter for a solenoid. | Computational Tools | |
| Calculate the number of conduction electrons. | Computational Tools | |
| Calculate the current in a circuit. | Computational Tools | |
| Calculate the terminal voltage of the battery. | Computational Tools | |
| Calculate resistivity at a specific temperature. | Computational Tools | |
| Calculate the coating thickness in meters. | Computational Tools | |
| Calculate new short-circuit current. | Computational Tools | |
| Calculate maximum power output (W). | Computational Tools | |
| Calculate the effective voltage after voltage drop. | Computational Tools | |
| Calculate the percentage of voltage drop. | Computational Tools | |
| Calculate the average current over a cycle. | Computational Tools | |
| Calculate the average voltage drop over a cycle. | Computational Tools | |
| Calculate the velocity of a charged particle in a uniform magnetic field. | Computational Tools | |
| Calculate the conductivity ratio assuming proportionality to mobility. | Computational Tools | |
| Calculate the new conductivity if original conductivity is known. | Computational Tools | |
| Calculate the power consumption of a resistor. | Computational Tools | |
| Assess whether the resistor’s rated power is sufficient and provide recommendation. | Computational Tools | |
| Determine the electric charge of a particle based on its type. | Computational Tools | |
| Calculate percentage change in resistance. | Computational Tools | |
| Calculate voltage per LSB for an ADC. | Computational Tools | |
| Convert inherent noise in LSB to voltage value. | Computational Tools | |
| Find the critical current density Jc0 at zero magnetic field. | Computational Tools | |
| Find the magnetic field strength H0 at zero critical current density. | Computational Tools | |
| Round the exact flux quantum number to an integer using specified method. | Computational Tools | |
| Calculate the new mobility after decrease. | Computational Tools | |
| Calculate the absolute decrease between two values. | Computational Tools | |
| Calculate the minimum magnetic field strength needed for magnetization. | Computational Tools | |
| Verify that the duty cycle is within the valid range [0, 1]. | Computational Tools | |
| Calculate heat released during combustion. | Computational Tools | |
| Calculate the net buoyancy coefficient from the density ratio. | Computational Tools | |
| Calculate the upward acceleration of the hot air balloon. | Computational Tools | |
| Calculate the ratio of target phonon mean free path to reference value. | Computational Tools | |
| Calculate the new welding speed after applying a speed factor. | Computational Tools | |
| Calculate the new laser power to maintain welding depth after increasing speed. | Computational Tools | |
| Calculate the theoretical welding depth. | Computational Tools | |
| Convert temperature from Celsius to Kelvin. | Computational Tools | |
| Calculate the thermal energy stored during a phase change. | Computational Tools | |
| Convert Celsius temperature to Kelvin. | Computational Tools | |
| Get water vapor pressure at a specific temperature. | Computational Tools | |
| Calculate the thermal resistance R = d / k. | Computational Tools | |
| Calculate the temperature difference. | Computational Tools | |
| Explain the relationship between thermal expansion coefficient ($`\alpha`$) and thermal strain rate sensitivity index (m). | Computational Tools | |
| Aggregate intermediate values and results into a dictionary. | Computational Tools | |
| Calculate heat absorbed. | Computational Tools | |
| Calculate the required heat energy. | Computational Tools | |
| Calculate the specific heat capacity (c) using heat energy, mass, and temperature change. | Computational Tools | |
| Return degeneracy array for each energy level. | Computational Tools | |
| Calculate the ratio of new energy to initial energy. | Computational Tools | |
| Calculate thermal conductivity of the target material from a known reference and ratio. | Computational Tools | |
| Generate a string for analytical temperature distribution expression. | Computational Tools | |
| Calculate the temperature difference. | Computational Tools | |
| Calculate the cooling time. | Computational Tools | |
| Calculate the temperature change. | Computational Tools | |
| Calculate the temperature gradient (°C/m) along a material. | Computational Tools | |
| Calculate the heat flux (W/m^2) using Fourier’s law. | Computational Tools | |
| Calculate the total heat conduction rate (W) through a material. | Computational Tools | |
| Calculate kinetic energy from energy density and volume. | Computational Tools | |
| Convert normalized temperature to Celsius. | Computational Tools | |
| Calculate temperature points T1 and T2 based on T_c and their ratios. | Computational Tools | |
| Calculate the energy released during phase change. | Computational Tools | |
| Calculate the gravitational potential energy. | Computational Tools | |
| Calculate the heat energy absorbed. | Computational Tools | |
| Calculate the temperature increase. | Computational Tools | |
| Calculate the difference between the thermal expansion coefficients of two materials. | Computational Tools | |
| Convert temperature to Kelvin. | Computational Tools | |
| Calculate Carnot efficiency. | Computational Tools | |
| Calculate the heat flux using Fourier’s law. | Computational Tools | |
| Calculate the total heat flow rate. | Computational Tools | |
| Calculate the total thermal resistance by summing individual resistances. | Computational Tools | |
| Add optional resistance to total if specified. | Computational Tools | |
| Calculate the heat energy required for heating. | Computational Tools | |
| Calculate the opposite (negation) of a heat value. | Computational Tools | |
| Calculate the mass of ice melted from the given energy. | Computational Tools | |
| Adjust energy to account for transfer efficiency. | Computational Tools | |
| Calculate the work done during an isothermal process for an ideal gas. | Computational Tools | |
| Calculate the change in internal energy for an ideal gas during an isothermal process. | Computational Tools | |
| Calculate the heat transfer based on the first law of thermodynamics. | Computational Tools | |
| Convert energy from MeV to Joules (J). | Computational Tools | |
| Calculate total heat based on enthalpy and moles. | Computational Tools | |
| Calculate heat per gram of adsorbent. | Computational Tools | |
| Calculate the temperature difference across a flat plate. | Computational Tools | |
| Return the Boltzmann constant (J/K). | Computational Tools | |
| Calculate heat generation rate per unit volume. | Computational Tools | |
| Calculate heat loss coefficient due to ventilation. | Computational Tools | |
| Calculate the final temperature in Kelvin. | Computational Tools | |
| Calculate time (seconds) to weld per millimeter. | Computational Tools | |
| Calculate heat input in Joules per millimeter. | Computational Tools | |
| Calculate the clearing temperature of a liquid crystal polymer. | Computational Tools | |
| Calculate dynamic viscosity $`\mu = \nu \cdot \rho`$. | Computational Tools | |
| Calculate the velocity gradient at $`y = -h`$ for the flow profile. | Computational Tools | |
| Calculate the drag coefficient (Cd) based on roughness. | Computational Tools | |
| Calculate the drag force based on fluid properties and drag coefficient. | Computational Tools | |
| Calculate total influent volume (m³/m²) over a given period. | Computational Tools | |
| Calculate the pore volume of the filter (m³/m²). | Computational Tools | |
| Calculate the fraction of filter pore volume occupied by solids. | Computational Tools | |
| Calculate infiltration through traditional pavement. | Computational Tools | |
| Calculate infiltration through permeable pavement. | Computational Tools | |
| Calculate the additional infiltration volume. | Computational Tools | |
| Calculate water mass in grams. | Computational Tools | |
| Compute the denominator for Jurin’s law: rho * g * d. | Computational Tools | |
| Calculate the capillary rise height. | Computational Tools | |
| Calculate the mass of the liquid. | Computational Tools | |
| Calculate the ratio of two densities. | Computational Tools | |
| Calculate gas permeability. | Computational Tools | |
| Calculate the minimum release height for a rolling solid sphere on a circular track. | Computational Tools | |
| Calculate incident photon rate. | Computational Tools | |
| Calculate the multiplicative effect of laser power increase on scattered photon rate. | Computational Tools | |
| Calculate reflectance at normal incidence using Fresnel equation. | Computational Tools | |
| Calculate the wavenumber for a hydrogen spectral line. | Computational Tools | |
| Calculate the wave number ratio k’/k in a dielectric material. | Computational Tools | |
| Calculate the wave number in a medium. | Computational Tools | |
| Calculate the transmission coefficient at the interface. | Computational Tools | |
| Calculate the minimum wavelength based on film thickness. | Computational Tools | |
| Calculate the maximum frequency for wave propagation. | Computational Tools | |
| Calculate the frequency range of electromagnetic waves. | Computational Tools | |
| Calculate the absorption ratio based on absorption coefficient and thickness. | Computational Tools | |
| Convert wavelength from nanometers to meters. | Computational Tools | |
| Generate wave function expressions for each quantum number. | Computational Tools | |
| Calculate photon energy in joules. | Computational Tools | |
| Determine if photon energy is sufficient to excite electron. | Computational Tools | |
| Calculate the numerical aperture (NA) of an optical fiber. | Computational Tools | |
| Calculate the emission rate W given free space rate and enhancement. | Computational Tools | |
| Calculate the rate of change of magnetic field. | Computational Tools | |
| Calculate the electron wavelength considering relativistic effects. | Computational Tools | |
| Calculate the Bragg angle in radians. | Computational Tools | |
| Calculate laser intensity. | Computational Tools | |
| Calculate radiation pressure. | Computational Tools | |
| Calculate photon energy in eV from wavelength. | Computational Tools | |
| Calculate total power in the spot area. | Computational Tools | |
| Calculate the photon flux per second. | Computational Tools | |
| Calculate the minimum film thickness for minimal normal incidence reflection. | Computational Tools | |
| Calculate the ratio of new irradiance to standard irradiance. | Computational Tools | |
| Calculate new max power point current. | Computational Tools | |
| Compute the difference between light and dark signal means. | Computational Tools | |
| Convert wavelength to meters. | Computational Tools | |
| Calculate volume in milliliters from mass and density. | Computational Tools | |
| Calculate the molar mass of a compound. | Computational Tools | |
| Convert mass to moles. | Computational Tools | |
| Calculate the mass of the unit cell based on density and volume. | Computational Tools | |
| Calculate number of atoms per unit cell. | Computational Tools | |
| Calculate volume in liters. | Computational Tools | |
| Calculate molarity of the solution. | Computational Tools | |
| Calculate pH from pOH. | Computational Tools | |
| Calculate the square root of the ratio of two molar masses. | Computational Tools | |
| Calculate the mass of solute in a saturated solution. | Computational Tools | |
| Calculate total mass of the saturated solution. | Computational Tools | |
| Calculate the mass percentage of the solute in the solution. | Computational Tools | |
| Calculate the reaction rate constant k from the half-life. | Computational Tools | |
| Calculate initial moles of gas using PV = nRT. | Computational Tools | |
| Calculate the number of molecules from moles. | Computational Tools | |
| Calculate remaining molecules after reaction_time_hours based on half-life. | Computational Tools | |
| Calculate the energy of an electron in a hydrogen atom at principal quantum number n. | Computational Tools | |
| Calculate the ionization energy as the absolute value of the energy level. | Computational Tools | |
| Calculate moles of hydrogen gas using ideal gas law. | Computational Tools | |
| Calculate the amount (moles) of gas. | Computational Tools | |
| Calculate the partial pressure of a gas (atm). | Computational Tools | |
| Calculate solution volume in mL from mass and density. | Computational Tools | |
| Determine the number of electrons transferred in the reaction. | Computational Tools | |
| Calculate Gibbs free energy change (J/mol). | Computational Tools | |
| Calculate the decay constant $`\lambda`$ given the known final mass, initial mass, and elapsed time. | Computational Tools | |
| Calculate the half-life $`T_{1/2}`$ from decay constant $`\lambda`$. | Computational Tools | |
| Calculate the remaining mass after a certain time using decay constant. | Computational Tools | |
| Calculate the mass of the content substance. | Computational Tools | |
| Calculate the molar amount of a component given total moles and mol%. | Computational Tools | |
| Retrieve the molar mass of a specified acid. | Computational Tools | |
| Convert density from g/mL to g/L. | Computational Tools | |
| Calculate the mass of acid in 1 liter of solution. | Computational Tools | |
| Calculate mass of a component based on weight fraction. | Computational Tools | |
| Convert weight percentage to weight fraction. | Computational Tools | |
| Calculate the final doping concentration after growth. | Computational Tools | |
| Return molecular mass in amu for a given molecule type. | Computational Tools | |
| Calculate the moles of Ni deposited. | Computational Tools | |
| Calculate the mass of Ni deposited in grams. | Computational Tools | |
| Calculate remaining moles of Ni ions after deposition. | Computational Tools | |
| Calculate the target moles of Ni ions after treatment. | Computational Tools | |
| Calculate the moles of Ni ions to be adsorbed. | Computational Tools | |
| Calculate the mass of adsorbent in kilograms. | Computational Tools | |
| Calculate the mass of the solute in grams. | Computational Tools | |
| Calculate $`Mg(OH)_2`$ solubility in pure water. | Computational Tools | |
| Calculate $`Mg(OH)_2`$ solubility in NaOH solution. | Computational Tools | |
| Calculate mass in grams from volume and density. | Computational Tools | |
| Calculate the required moles of calcium hydroxide based on acetic acid moles. | Computational Tools | |
| Convert concentration from mg/L to g/m³. | Computational Tools | |
| Calculate decay constant $`\lambda`$ from half-life. | Computational Tools | |
| Calculate time t from decay constant and activity ratio. | Computational Tools | |
| Calculate the total moles of a substance. | Computational Tools | |
| Calculate total number of atoms from moles. | Computational Tools | |
| Calculate the number of nanoparticles. | Computational Tools | |
| Calculate the total moles of nanoparticles. | Computational Tools | |
| Calculate the molar concentration of nanoparticles. | Computational Tools | |
| Calculate the scaling factor based on actual and reference PVC masses. | Computational Tools | |
| Calculate the additive mass needed based on the scaling factor. | Computational Tools | |
| Calculate the moles of CO2 produced from propane. | Computational Tools | |
| Calculate the bond order given bonding and antibonding electrons. | Computational Tools | |
| Estimate the bond length for a given bond type. | Computational Tools | |
| Generate an analysis statement relating bond order to bond length. | Computational Tools | |
| Calculate the curing time needed to reach a target degree of curing. | Computational Tools | |
| Calculate the degree of curing at time t. | Computational Tools | |
| Calculate corrosion rate. | Computational Tools | |
| Calculate the number of unit cells in one mole of the compound. | Computational Tools | |
| Calculate the total volume occupied by one mole of the compound. | Computational Tools | |
| Calculate the molar mass of the compound. | Computational Tools | |
| Calculate the mass of a component. | Computational Tools | |
| Calculate the mass of each oxide. | Computational Tools | |
| Calculate zinc and copper masses from total mass and zinc percentage. | Computational Tools | |
| Calculate the number of atoms from moles. | Computational Tools | |
| Calculate total gas molecules based on volume and molecules per cc. | Computational Tools | |
| Calculate the atomic radius in FCC structure. | Computational Tools | |
| Calculate twice the atomic radius. | Computational Tools | |
| Calculate the crystallinity percentage based on density measurements. | Computational Tools | |
| Calculate the plating time needed to reach the target thickness. | Computational Tools | |
| Calculate alcohol proof from volume percentage. | Computational Tools | |
| Calculate required $`LiFePO_4`$ cathode material mass. | Computational Tools | |
| Calculate the number of atoms from moles. | Computational Tools | |
| Determine the number of atoms per unit cell based on crystal structure. | Computational Tools | |
| Calculate the mass of sodium benzoate. | Computational Tools | |
| Calculate the nucleophilic Fukui function $`f^{+}`$. | Computational Tools | |
| Calculate the electrophilic Fukui function $`f^{-}`$. | Computational Tools | |
| Calculate the Dual Descriptor f. | Computational Tools | |
| Return detailed explanation of the physical meaning of adsorption enthalpy ($`\Delta H_{ads}`$). | Computational Tools | |
| Calculate total coordination number based on cyclic ligand denticity and additional ligands. | Computational Tools | |
| Return the typical coordination number range for lanthanide elements. | Computational Tools | |
| Assess whether the total coordination number is within the typical lanthanide range. | Computational Tools | |
| Calculate the sum of two ionic radii, representing the distance between ions. | Computational Tools | |
| Calculate the tolerance factor t. | Computational Tools | |
| Assess perovskite structure stability based on tolerance factor t. | Computational Tools | |
| Parse electron configuration string, extract number of d electrons. | Computational Tools | |
| Calculate expected d electron count based on oxidation state, for verification. | Computational Tools | |
| Calculate total spin angular momentum S from number of d electrons. | Computational Tools | |
| Calculate the standard cell potential (E°) from cathode and anode potentials. | Computational Tools | |
| Calculate the reaction quotient Q for the cell reaction. | Computational Tools | |
| Calculate the mass of a single suppository. | Computational Tools | |
| Return gas constant R in J/(mol·K). | Computational Tools | |
| Return Faraday constant F in C/mol. | Computational Tools | |
| Calculate the coefficient (RT)/(nF) for the Nernst equation. | Computational Tools | |
| Generate a sequence of molecular weight values within a specified range. | Computational Tools | |
| Calculate total mass of suspended solids in grams. | Computational Tools | |
| Calculate the final density of a material. | Computational Tools | |
| Calculate total PEG mass needed for all suppositories. | Computational Tools | |
| Calculate the sum of length and width given the perimeter. | Computational Tools | |
| Calculate the product of length and width given the volume and height. | Computational Tools | |
| Ensure that length >= width in the dimensions tuple. | Computational Tools | |
| Select the first dimension set as the final dimensions. | Computational Tools | |
| Round a value to a specified number of decimal places. | Computational Tools | |
| Calculate the area of a rectangle. | Computational Tools | |
| Calculate the angle $`\Theta`$ in radians from shear strain. | Computational Tools | |
| Convert radians to degrees. | Computational Tools | |
| Calculate $`\Phi`$ in degrees from $`\Theta`$ in degrees. | Computational Tools | |
| Calculate cot(2$`\Phi`$) as tan($`\Theta`$). | Computational Tools | |
| Verify if cot(2$`\Phi`$) is close to shear_strain / 2. | Computational Tools | |
| Calculate the volume of a rectangular prism. | Computational Tools | |
| Calculate volume in cubic centimeters. | Computational Tools | |
| Round a float to the nearest integer. | Computational Tools | |
| Calculate the area of a curb segment. | Computational Tools | |
| Sum a list of area values. | Computational Tools | |
| Round the volume in cubic feet to a specified number of decimal places. | Computational Tools | |
| Calculate the cross-sectional area of a rectangular prism. | Computational Tools | |
| Return the denominator (plate separation d). | Computational Tools | |
| Calculate the change in length. | Computational Tools | |
| Round a number to a specified number of significant figures. | Computational Tools | |
| Derive the gamma distribution’s MGF symbolically using sympy. | Computational Tools | |
| Round a value to a specified number of decimal places. | Computational Tools | |
| Calculate the diameter of a sphere given its radius. | Computational Tools | |
| Calculate total volume of all atoms in the unit cell. | Computational Tools | |
| Calculate total volume from component volumes. | Computational Tools | |
| Calculate volume fraction of a component. | Computational Tools | |
| Calculate the volume of a sphere given its radius. | Computational Tools | |
| Generate mesh coordinates for a sphere surface. | Computational Tools | |
| Calculate cross-sectional area. | Computational Tools | |
| Calculate the increase factor of K-points. | Computational Tools | |
| Calculate the percentage decrease based on absolute decrease and initial value. | Computational Tools | |
| Convert angle from degrees to radians. | Computational Tools | |
| Convert a percentage to a decimal ratio. | Computational Tools | |
| Compute the ratio of numerator to denominator. | Computational Tools | |
| Calculate the volume of a hollow cylinder. | Computational Tools | |
| Compute the constant c analytically so that the integral of f(x) = cx^2 over [0,1] equals 1. | Computational Tools | |
| Convert a percentage value to decimal. | Computational Tools | |
| Calculate the cosine of an angle in radians. | Computational Tools | |
| Calculate the ratio of initial to final values. | Computational Tools | |
| Calculate the sine of an angle in radians. | Computational Tools | |
| Calculate the height of the ramp. | Computational Tools | |
| Round a number to the nearest tenth. | Computational Tools | |
| Calculate the absolute difference between two numbers. | Computational Tools | |
| Calculate the number of bags needed to cover an area. | Computational Tools | |
| Calculate the total area of a rectangular region. | Computational Tools | |
| Calculate the area to be seeded with grass. | Computational Tools | |
| Calculate the area of a rectangle. | Computational Tools | |
| Calculate the volume of a unit cell. | Computational Tools | |
| Convert a ratio to a percentage. | Computational Tools | |
| Calculate the percentage increase from original_value to new_value. | Computational Tools | |
| Calculate maximum number of items that can fit into the storage space. | Computational Tools | |
| Calculate maximum storage days based on volume and space utilization. | Computational Tools | |
| Calculate the face diagonal length of the cubic cell. | Computational Tools | |
| Calculate the distance between neighboring atom centers. | Computational Tools | |
| Return the original algebraic expression string for the difference in tank volumes. | Computational Tools | |
| Factorize the formula for the volume difference. | Computational Tools | |
| Validate the correctness of the factorization. | Computational Tools | |
| Calculate z-coordinate of the curve at parameter t. | Computational Tools | |
| Calculate dz/dt at parameter t. | Computational Tools | |
| Calculate the analytical probability that X < value. | Computational Tools | |
| Convert angle from degrees to radians. | Computational Tools | |
| Calculate the square root of the grain size. | Computational Tools | |
| Calculate the area of a square. | Computational Tools | |
| Calculate the minimum velocity at the top of the track. | Computational Tools | |
| Format the volume to a string with two decimal places. | Computational Tools | |
| Estimate total orbital angular momentum L, simplified. | Computational Tools | |
| Convert a set to a sorted list. | Computational Tools | |
| Calculate the surface area of a cube-shaped warehouse. | Computational Tools | |
| Validate that the sum of squared Miller indices is not zero. | Computational Tools | |
| Compute the sum of squares of Miller indices. | Computational Tools | |
| Calculates the symbolic expectation E[X] of the distribution using sympy. | Computational Tools | |
| Calculates the symbolic second moment E[X^2] using sympy. | Computational Tools | |
| Calculate the initial area of the window. | Computational Tools | |
| Convert diameter (millimeter) to radius (meter). | Computational Tools | |
| Convert wire diameter to radius. | Computational Tools | |
| Calculate the length contraction due to thermal contraction. | Computational Tools | |
| Calculate the final volume after volume reduction. | Computational Tools | |
| Calculate total manufacturing overhead costs. | Computational Tools | |
| Calculate direct manufacturing costs. | Computational Tools | |
| Calculate total manufacturing costs. | Computational Tools | |
| Calculate non-manufacturing costs. | Computational Tools | |
| Calculate total costs. | Computational Tools | |
| Calculate gross profit. | Computational Tools | |
| Calculate net profit. | Computational Tools | |
| Calculate total input cost from labor, material, and overhead costs. | Computational Tools | |
| Calculate productivity ratio as output value divided by total input cost. | Computational Tools | |
| Calculate explicit costs. | Computational Tools | |
| Calculate opportunity costs. | Computational Tools | |
| Calculate accounting profit. | Computational Tools | |
| Calculate economic profit. | Computational Tools | |
| Calculate total units to account for. | Computational Tools | |
| Calculate units completed and transferred out. | Computational Tools | |
| Calculate the indirect materials cost. | Computational Tools | |
| Calculate the indirect labor cost. | Computational Tools | |
| Calculate total direct costs. | Computational Tools | |
| Calculate individual element’s contribution to total magnetic moment. | Computational Tools | |
| Calculate total materials used by summing direct and indirect materials. | Computational Tools | |
| Calculate ending inventory of raw materials. | Computational Tools | |
| Calculate the absolute error between measured value and true value. | Computational Tools | |
| Calculate the percentage error. | Computational Tools | |
| Analyze possible causes of measurement error. | Computational Tools | |
| Convert input data to a NumPy array. | Computational Tools | |
| Convert a number to scientific notation with specified significant digits. | Computational Tools | |
| Format mantissa and exponent into a scientific notation string. | Computational Tools | |
| Calculate the absolute error between measured and true values. | Computational Tools | |
| Calculate the magnitude (absolute value) of an error. | Computational Tools | |
| Calculate the relative error as a percentage. | Computational Tools | |
| Round a value to a specified number of decimal places. | Computational Tools | |
| Calculate sample variance from Sxx and sample size. | Computational Tools | |
| Format output pore volume and unit. | Computational Tools | |
| Calculate percentage change from relative change ratio. | Computational Tools | |
| Calculate the percentage error based on absolute difference and reference value. | Computational Tools | |
| Calculate the mean square (MS) from sum of squares (SS) and degrees of freedom (DF). | Computational Tools | |
| Calculate the F-value for an ANOVA test. | Computational Tools | |
| Compute the critical F value for given significance level and degrees of freedom. | Computational Tools | |
| Determine if the F value indicates significance. | Computational Tools | |
| Calculate the maximum integer value for a given bit resolution. | Computational Tools | |
| Convert normalized humidity to percentage. | Computational Tools | |
| Calculate the relative uncertainty. | Computational Tools | |
| Calculate the combined relative uncertainty using root-sum-square. | Computational Tools | |
| Calculate the relative error as a ratio. | Computational Tools | |
| Check if two arrays have the same length. | Computational Tools | |
| Print the input parameters with formatting for clarity. | Computational Tools | |
| Calculate extra space needed if storage is insufficient. | Computational Tools | |
| Convert a ratio to scientific notation with mantissa rounded to one decimal. | Computational Tools | |
| Calculate the probability density function at point(s) x. | Computational Tools | |
| Convert input to a numpy array. | Computational Tools | |
| Generate a list of integer q values from start to end inclusive. | Computational Tools | |
| Calculate the theoretical variance of a normal distribution. | Computational Tools | |
| Calculate the median of a list of numerical data. | Computational Tools | |
| Calculate the mode(s) of a list of numerical data. | Computational Tools | |
| Convert an input iterable to a set. | Computational Tools | |
| Calculate the maximum quantization error, equal to half the LSB voltage. | Computational Tools | |
| Compute the difference between light and dark signal variances. | Computational Tools | |
| Calculate the standard deviation from the variance difference. | Computational Tools | |
| Validate that interval individuals do not exceed total individuals. | Computational Tools | |
| Compute interval relative density. | Computational Tools | |
| Calculate probability that device life exceeds threshold. | Computational Tools | |
| Create mesh grids for electron density and temperature ranges. | Computational Tools | |
| Convert length from millimeters to meters. | Computational Tools | |
| Convert volume from milliliters to deciliters. | Computational Tools | |
| Convert current from milliamperes to amperes. | Computational Tools | |
| Convert length from picometers to centimeters. | Computational Tools | |
| Convert capacitance from Farads to specified units. | Computational Tools | |
| Convert weight from kilograms to tons. | Computational Tools | |
| Convert mass from kilograms to grams. | Computational Tools | |
| Convert pressure from mm mercury to atmospheres. | Computational Tools | |
| Convert volume from cubic meters to cubic feet. | Computational Tools | |
| Convert length from kilometers to meters. | Computational Tools | |
| Convert length from centimeters to meters. | Computational Tools | |
| Convert pressure from mm Hg to atm. | Computational Tools | |
| Convert mass to grams. | Computational Tools | |
| Convert volume to milliliters. | Computational Tools | |
| Convert thickness from nanometers to meters. | Computational Tools | |
| Convert film thickness to meters. | Computational Tools | |
| Convert area from square centimeters to square meters. | Computational Tools | |
| Convert cross-sectional area from mm² to m². | Computational Tools | |
| Convert length from millimeters to meters. | Computational Tools | |
| Convert force from kilonewtons to newtons. | Computational Tools | |
| Convert mass from kilograms to grams. | Computational Tools | |
| Convert grain size from micrometers to meters. | Computational Tools | |
| Convert length from nanometers to meters. | Computational Tools | |
| Convert energy from Joules to electron volts. | Computational Tools | |
| Convert area to square meters based on unit system. | Computational Tools | |
| Convert mass from grams to kilograms. | Computational Tools | |
| Convert volume from liters to cubic meters. | Computational Tools | |
| Convert time from minutes to seconds. | Computational Tools | |
| Convert thickness from nanometers to micrometers. | Computational Tools | |
| Convert force from picoNewtons to Newtons. | Computational Tools | |
| Convert distance to meters if input is in centimeters. | Computational Tools | |
| Convert length change from meters to millimeters. | Computational Tools | |
| Convert velocity from kilometers per hour to meters per second. | Computational Tools | |
| Convert atomic mass units to kilograms. | Computational Tools | |
| Convert volume from liters to milliliters. | Computational Tools | |
| Convert mass from grams to kilograms. | Computational Tools | |
| Convert mass from grams to pounds. | Computational Tools | |
| Convert work function to Joules. | Computational Tools | |
| Format the density unit string. | Computational Tools | |
| Convert pressure from Torr to atm. | Computational Tools | |
| Convert energy to Joules. | Computational Tools | |
| Convert dimension to meters. | Computational Tools | |
| Convert time in hours to days. | Computational Tools | |
| Convert radius from centimeters to meters if necessary. | Computational Tools | |
| Convert length from inches to centimeters. | Computational Tools | |
| Convert length from meters to centimeters. | Computational Tools | |
| Convert length from meters to millimeters. | Computational Tools | |
| Convert volume from milliliters to cubic meters. | Computational Tools | |
| Convert inductance from millihenries to henries. | Computational Tools | |
| Convert H2 production rate from mmol/cm²/h to molecules/cm²/s. | Computational Tools | |
| Convert length from meters to specified units. | Computational Tools | |
| Convert length to meters. | Computational Tools | |
| Convert area to square meters. | Computational Tools | |
| Convert length from meters to target units. | Computational Tools | |
| Convert work function from electron volts to joules. | Computational Tools | |
| Convert force from kilonewtons to newtons. | Computational Tools | |
| Convert volume from liters to cubic centimeters. | Computational Tools | |
| Convert thickness from millimeters to micrometers. | Computational Tools | |
| Convert elastic modulus from GPa to MPa. | Computational Tools | |
| Convert energy units based on a conversion factor. | Computational Tools | |
| Convert speed from km/h to m/s. | Computational Tools | |
| Convert thickness from kilometers to meters. | Computational Tools | |
| Convert carrier concentration from cm^-3 to m^-3. | Computational Tools | |
| Convert mobility from cm^2/(V·s) to m^2/(V·s). | Computational Tools | |
| Convert volume to cubic centimeters based on the input unit. | Computational Tools | |
| Convert density from g/cm³ to g/m³. | Computational Tools | |
| Convert volume from liters to cubic centimeters. | Computational Tools | |
| Convert length from meters to micrometers. | Computational Tools | |
| Convert gallons to liters. | Computational Tools | |
| Convert milliliters to cubic centimeters. | Computational Tools | |
| Convert microcuries ($`\mu`$Ci) to becquerel (Bq). | Computational Tools | |
| Convert maximum power from watts to kilowatts. | Computational Tools | |
| Convert energy from electron volts to joules. | Computational Tools | |
| Convert radius from meters to centimeters. | Computational Tools | |
| Convert distance from mm to m. | Computational Tools | |
| Convert pressure from kPa to Pa. | Computational Tools | |
| Convert bulk modulus from kPa to Pa. | Computational Tools | |
| Convert stress from pascal (Pa) to megapascal (MPa). | Computational Tools | |
| Convert welding speed from m/min to mm/s. | Computational Tools | |
| Convert length from centimeters to millimeters, rounded to 2 decimal places. | Computational Tools | |
| Return a dictionary of fundamental physical constants. | Computational Tools | |
| Complete structure analysis from compound name (SMILES + properties). | Computational Tools | |
| Calculate molecular weight from compound name. | Computational Tools | |
| Identify all functional groups in a molecule. | Computational Tools | |
| Calculate comprehensive molecular descriptors. | Computational Tools | |
| Check Lipinski’s Rule of Five for drug-likeness. | Computational Tools | |
| Compare two molecules for similarity using Tanimoto coefficient. | Computational Tools | |
| Generate multiple types of molecular fingerprints. | Computational Tools | |
| Analyze stereochemical properties. | Computational Tools | |
| Analyze ring systems in molecules. | Computational Tools | |
| Analyze 3D conformational properties. | Computational Tools | |
| Calculate electronic properties of molecules. | Computational Tools | |
| Calculate molecular shape descriptors. | Computational Tools | |
| Comprehensive chirality analysis. | Computational Tools | |
| Analyze bond properties in molecules. | Computational Tools | |
| Analyze heteroatom content. | Computational Tools | |
| Calculate formal charges. | Computational Tools | |
| Analyze molecular fragments. | Computational Tools | |
| Calculate topological indices. | Computational Tools | |
| Calculate molecular kappa shape indices. | Computational Tools | |
| Analyze aromatic systems. | Computational Tools | |
| Convert between molecular structure formats. | Computational Tools | |
| Convert compound name to all structure formats. | Computational Tools | |
| Resolve InChIKey to other formats. | Computational Tools | |
| Bidirectional SELFIES conversion. | Computational Tools | |
| Validate and standardize molecular structures. | Computational Tools | |
| Generate tautomers of a molecule. | Computational Tools | |
| Validate InChI and InChIKey strings. | Computational Tools | |
| Standardize molecular structures. | Computational Tools | |
| Search for substructures in molecules. | Computational Tools | |
| Calculate comprehensive protein properties. | Computational Tools | |
| Analyze protein stability indicators. | Computational Tools | |
| Perform sequence alignment analysis. | Computational Tools | |
| Find common protein motifs. | Computational Tools | |
| Predict protein solubility. | Computational Tools | |
| Analyze antibody sequence features. | Computational Tools | |
| Predict subcellular localization. | Computational Tools | |
| Analyze amino acid composition. | Computational Tools | |
| Predict protein-protein interaction potential. | Computational Tools | |
| Predict drug-target interaction. | Computational Tools | |
| Comprehensive DNA sequence analysis. | Computational Tools | |
| Translate DNA to protein. | Computational Tools | |
| Find DNA complement and reverse complement. | Computational Tools | |
| Find palindromic sequences. | Computational Tools | |
| Optimize codon usage. | Computational Tools | |
| Optimize DNA/RNA codons. | Computational Tools | |
| Design PCR primers. | Computational Tools | |
| Analyze restriction enzyme sites. | Computational Tools | |
| Analyze circular DNA. | Computational Tools | |
| Generate random DNA sequence. | Computational Tools | |
| Calculate peptide properties. | Computational Tools | |
| Design alanine scanning library. | Computational Tools | |
| Design truncation library. | Computational Tools | |
| Design overlapping peptide library. | Computational Tools | |
| Design positional scanning library. | Computational Tools | |
| Analyze protease digestion patterns. | Computational Tools | |
| Calculate degenerate codons. | Computational Tools | |
| Calculate oligonucleotide properties. | Computational Tools | |
| Convert peptide sequence to SMILES. | Computational Tools | |
| Compute binding affinity. | Computational Tools | |
| Predict ADMET properties for compounds. | Computational Tools | |
| Analyze drug-likeness properties. | Computational Tools | |
| Predict blood-brain barrier penetrance. | Computational Tools | |
| Predict oral bioavailability. | Computational Tools | |
| Convert CID to comprehensive properties. | Computational Tools | |
| Optimize molecule for drug-likeness. | Computational Tools | |
| Analyze lead compound for optimization. | Computational Tools | |
| Analyze molecular scaffold. | Computational Tools | |
| Generate random molecules. | Computational Tools | |
| Enumerate all tautomers. | Computational Tools | |
| Predict protein-ligand interactions. | Computational Tools | |
| Complete target protein profile. | Computational Tools | |
| Validate drug-target pair. | Computational Tools | |
| Calculate small molecule similarity. | Computational Tools | |
| Analyze MOF structure. | Computational Tools | |
| Calculate material density. | Computational Tools | |
| Analyze material symmetry. | Computational Tools | |
| Analyze element composition. | Computational Tools | |
| Convert MOF to SMILES. | Computational Tools | |
| Analyze MOF lattice parameters. | Computational Tools | |
| Complete material analysis. | Computational Tools | |
| Convert SMILES to CAS number. | Computational Tools | |
| Analyze small molecule-protein interaction. | Computational Tools | |
| Design peptide-based drugs. | Computational Tools | |
| Screen bioactive compounds. | Computational Tools | |
| Convert peptide to SMILES and analyze. | Computational Tools | |
| Optimize peptide for drug properties. | Computational Tools | |
| Complete compound ADMET profile. | Computational Tools | |
| Screen drug candidates. | Computational Tools | |
| Profile lead compound. | Computational Tools | |
| Score compound drug-likeness. | Computational Tools | |
| Analyze drug similarity. | Computational Tools | |
| Design drugs for protein target. | Computational Tools | |
| Design antibody-drug conjugates. | Computational Tools | |
| Analyze biologic drugs. | Computational Tools | |
| Validate target sequence for druggability. | Computational Tools | |
| Profile protein-drug interactions. | Computational Tools | |
| Analyze MOF as compound. | Computational Tools | |
| Convert material to compound format. | Computational Tools | |
| Calculate MOF properties. | Computational Tools | |
| Analyze crystal structure. | Computational Tools | |
| Analyze material for drug delivery. | Computational Tools | |
| Compute various physical and chemical parameters for a given protein sequence using Expasy ProtParam API. | Computational Tools | |
| Predict the hydrophilicity of a protein sequence. | Computational Tools | |
| This tool compute the molar extinction coefficient and protein concentration of the protein, and also provides information such as the protein isoelectric point. | Computational Tools | |
| Compute the theoretical isoelectric point (pI) and molecular weight (mW) of a protein sequence. | Computational Tools | |
| Codon Optimization Tool: Used to optimize codons for expression of recombinant genes in mainstream hosts. | Computational Tools | |
| Peptide Property Calculator: Calculate molecular weight, extinction coefficient, net peptide charge, peptide isoelectric point, and average hydrophobicity (GRAVY) of peptide properties. | Computational Tools | |
| Oligonucleotide (primer) Calculator: The annealing temperature (Tm), molecular weight (MW), extinction coefficient (OD/$`\mu`$mol, $`\mu`$g/OD) of the oligonucleotides were calculated. | Computational Tools | |
| This tool optimize the the expression of recombinant gene condons of the protein, Input: protein: protein sequence Returns: str: The Markdown content with new sequence. | Computational Tools | |
| This tool optimize the the expression of recombinant gene condons of the DNA/RNA, Input: protein: DNA or RNA sequence. | Computational Tools | |
| This tool compute the hydrophilicity of the protein, | Computational Tools | |
| This tool compute the annealing temperature of an oligonucleotide. | Computational Tools | |
| This tool translate the polypeptide sequence to SMILES. | Computational Tools | |
| This tool compute the isoelectric point of the protein or peptide. | Computational Tools | |
| This tool compute affinity based on the molar Gibbs free energy | Computational Tools | |
| This tool compute the Average molecular weight of the polypeptide. | Computational Tools | |
| This tool compute the chemical formula of the polypeptide. | Computational Tools | |
| This tool calculates the optimal degenerate codons that encode one or more input amino acids. | Computational Tools | |
| This tool design overlapping peptide library. | Computational Tools | |
| This tool design peptide library. | Computational Tools | |
| This tool design truncation peptide library. | Computational Tools | |
| This tool design positional scanning peptide library. | Computational Tools | |
| This tool can simulate the hydrolytic behavior of protein-degrading enzymes. | Computational Tools | |
| This tool label the variable regions of antibodies with CDR and FR regions; Users need to choose a numbering system, and the numbering schemes include: imgt, chothia, kabat, martin; The definition scheme includes chothia, kabat, imgt, and contact. | Computational Tools | |
| This tool number the amino acid sequence of the antibody; Identify the input sequence and distinguish between immunoglobulin (IG) and T cell receptor (TR); The numbering system includes: IMGT, Chothia, Kabat, Martin (extended version Chothia), and AHo; TR sequences can only be numbered using IMGT or AHo. | Computational Tools | |
| This tool aligns circular DNA sequences. | Computational Tools | |
| Multiple Sequence Comparison by Log Expectation is a tool used to compare protein or nucleic acid sequences. | Computational Tools | |
| This tool compares two sequences in global alignment style. | Computational Tools | |
| This tool predict the inherent disordered regions of the protein based on sequence. | Computational Tools | |
| This tool compare two sequences in local alignment style. | Computational Tools | |
| This tool analyse the motif of the protein based on sequence. | Computational Tools | |
| Sequence similarity calculation takes a set of aligned sequences (FASTA or GCG format) as input to calculate their similarity | Computational Tools | |
| The ORF search tool can help you find open reading frames in DNA sequences, and the returned results include the start and end positions of the ORF as well as the translation results of the open reading frames. | Computational Tools | |
| This tool translate DNA sequence to protein(amino acid) sequence. | Computational Tools | |
| This tool search repeat DNA sequence in DNA sequence. | Computational Tools | |
| This tool search repeat protein sequence in DNA sequence. | Computational Tools | |
| This tool searches for palindrome sequences in the sequence and enters the length range of nucleic acid sequences and palindrome sequences in the text box below. | Computational Tools | |
| This tool calculate amino acid by degenerate codon. | Computational Tools | |
| This tool predict nuclear localization sequence of protein based on sequence. | Computational Tools | |
| This tool calculate the similarity of small molecules based on SMILES. | Computational Tools | |
| This tool calculate the molecular weight of DNA based on sequence. | Computational Tools | |
| The CpG island prediction tool can predict potential CpG islands using the Gardiner Garden and Frommer (1987) method The calculation method is to use a 200bp window, with each shift of 1 bp. | Computational Tools | |
| This tool calculate the properties of PCR primer based on sequence. | Computational Tools | |
| This tool count the number of amino acids in the protein. | Computational Tools | |
| The enzyme digestion site summary tool counts the number and location of commonly used restriction endonucleating recognition sites in DNA sequences. | Computational Tools | |
| This tool generate random DNA sequence. | Computational Tools | |
| Calculates the molecular weight of a structure in a PDB file using MDAnalysis. | Computational Tools | |
| Calculates the frequency of each amino acid in a protein sequence. | Computational Tools | |
| Generates the reverse complement of a DNA sequence. | Computational Tools | |
| Calculates the hydrophobicity and polarity of a protein sequence. | Computational Tools | |
| Convert multiple MOF materials into SMILES representations and return the results in Markdown table format. | Computational Tools | |
| Reads a structure file and returns basic information about the structure. | Computational Tools | |
| Calculates the density of a structure from a file. | Computational Tools | |
| Returns the elemental composition of a structure from a file. | Computational Tools | |
| Calculates the symmetry of a structure from a file. | Computational Tools | |
| Identify and list the functional groups in a molecule given its SMILES string. | Computational Tools | |
| Calculate the molecular weight of a molecule given its SMILES string. | Computational Tools | |
| Calculate the Tanimoto similarity between two molecules given their SMILES strings. | Computational Tools | |
| Convert a SMILES string to an InChI string. | Computational Tools | |
| Convert an InChIKey string to a SMILES string. | Computational Tools | |
| Convert an InChIKey string to InChI. | Computational Tools | |
| Convert an InChI string to a MOL string. | Computational Tools | |
| Check if an InChIKey string is valid. | Computational Tools | |
| Convert an InChI string to a SMILES string. | Computational Tools | |
| Convert an InChI string to an InChIKey string. | Computational Tools | |
| Convert InChI to ChemSpider ID. | Computational Tools | |
| Translates a SMILES string into its corresponding SELFIES string. | Computational Tools | |
| Translates a SELFIES string into its corresponding SMILES string. | Computational Tools | |
| Generates a random molecule. | Computational Tools | |
| Computes the length of a SELFIES string. | Computational Tools | |
| Splits a SELFIES string into its individual tokens. | Computational Tools | |
| Generate the atom pair fingerprint of a molecule as a SparseBitVect. | Computational Tools | |
| Assign Patty types to the atoms of a molecule. | Computational Tools | |
| Perform a series of tests on a molecule, including sanitization, removal of hydrogens, and canonicalization check. | Computational Tools | |
| Generate a molecule image from its SMILES representation and embed it directly in Markdown. | Computational Tools | |
| Assigns EState types to each atom in a molecule based on its SMILES representation. | Computational Tools | |
| Calculate EState indices for each atom in a molecule based on its SMILES representation. | Computational Tools | |
| Calculate EState VSA indices for a molecule based on its SMILES representation. | Computational Tools | |
| Generate the EState fingerprint for a molecule based on its SMILES representation. | Computational Tools | |
| Calculate shape similarity scores using USRCAT for a list of molecules defined by their SMILES. | Computational Tools | |
| Calculate the normalized principal moments of inertia (NPR1 and NPR2) for a molecule. | Computational Tools | |
| Calculate the distance matrix for a list of molecules based on their fingerprints. | Computational Tools | |
| Clusters molecules based on their fingerprints and returns the clustering results in Markdown format. | Computational Tools | |
| Process the molecular fingerprint generated by FingerprintMol function. | Computational Tools | |
| Generate fingerprints for a list of SMILES strings. | Computational Tools | |
| Generate an RDKit fingerprint from a SMILES string using default parameters. | Computational Tools | |
| Generate all possible Fraggle fragmentations for a molecule represented by a SMILES string. | Computational Tools | |
| Check if the molecule represented by a SMILES string is a valid ring cut. | Computational Tools | |
| Generate an Atom Pair Fingerprint from a SMILES string and display the results in a readable format. | Computational Tools | |
| Generate a Torsions Fingerprint from a SMILES string. | Computational Tools | |
| Generate an RDKit fingerprint from a SMILES string. | Computational Tools | |
| Generate a Pharm2D fingerprint from a SMILES string. | Computational Tools | |
| Generate a Morgan fingerprint from a SMILES string. | Computational Tools | |
| Generate an Avalon fingerprint from a SMILES string. | Computational Tools | |
| Converts a SMILES string to its corresponding InChI string. | Computational Tools | |
| Generates a molecular key for a given molecule represented by a SMILES string. | Computational Tools | |
| Generates the stereo code for a given molecule represented by a SMILES string. | Computational Tools | |
| The tool is used to determine the bond orders between atoms in a molecule based on their atomic coordinates. | Computational Tools | |
| The tool is used to determine the bond orders between atoms in a molecule based on their atomic coordinates. | Computational Tools | |
| This tool is used to generate a pattern fingerprint for a molecule. | Computational Tools | |
| This tool is used to check if a molecule(target) is a substructure of another molecule(template). | Computational Tools | |
| This tool is used to get the template molecule from a TautomerQuery object. | Computational Tools | |
| This tool obtains all possible tautomers of a TautomerQuery object. | Computational Tools | |
| This tool is used to get the modified atoms of a TautomerQuery object. | Computational Tools | |
| This tool is used to get the modified bonds of a TautomerQuery object. | Computational Tools | |
| This tool is to search for substructures in a given target molecule that match the tautomer query. | Computational Tools | |
| This tool is used to check if a TautomerQuery object can be serialized. | Computational Tools | |
| This tool is used to assign CIP labels to the atoms in a molecule. | Computational Tools | |
| The rdkit. | Computational Tools | |
| The rdkit. | Computational Tools | |
| This tool finds and replaces abbreviation substance groups in a molecule, resulting in a compressed version of the molecule where the abbreviations are expanded. | Computational Tools | |
| This tool is used to convert a SLN string to a SMILES string. | Computational Tools | |
| This tool is used to create a shingling for a molecule. | Computational Tools | |
| This tool creates an MHFP vector from a molecule using MHFP encoder, capturing structural information of the molecule. | Computational Tools | |
| This tool creates an SECFP vector from a molecule using SECFP encoder, capturing structural information of the molecule. | Computational Tools | |
| This tool computes the 2D BCUT descriptors for a given molecule, representing mass, Gasteiger charge, Crippen logP, and Crippen MR values. | Computational Tools | |
| This tool computes the 2D autocorrelation descriptors for a given molecule, capturing the spatial arrangement of atoms in the molecule. | Computational Tools | |
| This tool computes the 3D autocorrelation descriptors for a given molecule, capturing the spatial arrangement of atoms in the molecule. | Computational Tools | |
| This tool calculates the asphericity descriptor for a molecule, which measures how much the molecule deviates from a perfectly spherical shape. | Computational Tools | |
| This tool calculates the chi^0 (chi-zero) cluster index, which represents a topological descriptor related to molecular branching. | Computational Tools | |
| This function calculates the Chi^0v (Chi-zero-v) valence molecular graph index for a molecule, which is used to describe the topology of the molecule. | Computational Tools | |
| This tool calculates the chi^1 (chi-one) cluster index, which represents a topological descriptor related to molecular branching. | Computational Tools | |
| This function calculates the Chi^1v (Chi-one-v) valence molecular graph index for a molecule, which is used to describe the topology of the molecule. | Computational Tools | |
| This tool calculates the chi^2 (chi-two) cluster index, which represents a topological descriptor related to molecular branching. | Computational Tools | |
| This function calculates the Chi^2v (Chi-two-v) valence molecular graph index for a molecule, which is used to describe the topology of the molecule. | Computational Tools | |
| This tool calculates the chi^3 (chi-three) cluster index, which represents a topological descriptor related to molecular branching. | Computational Tools | |
| This function calculates the Chi^3v (Chi-three-v) valence molecular graph index for a molecule, which is used to describe the topology of the molecule. | Computational Tools | |
| This tool calculates the chi^4 (chi-four) cluster index, which represents a topological descriptor related to molecular branching. | Computational Tools | |
| This function calculates the Chi^4v (Chi-four-v) valence molecular graph index for a molecule, which is used to describe the topology of the molecule. | Computational Tools | |
| This tool calculates the Coulomb matrix for a molecule, which represents the electrostatic interactions between atoms in the molecule. | Computational Tools | |
| This function calculates the Wildman-Crippen logP and MR (molecular refractivity) values for a given molecule in RDKit. | Computational Tools | |
| This function computes the EEM (Electronegativity Equalization Method) atomic partial charges for a given molecule using its atomic properties. | Computational Tools | |
| This function calculates the eccentricity of a molecule, which is a measure of its shape. | Computational Tools | |
| This function calculates the exact molecular weight of a molecule, which is the sum of the atomic weights of all atoms in the molecule. | Computational Tools | |
| This function calculates the fraction of sp3-hybridized carbon atoms in a molecule, which is a measure of its shape. | Computational Tools | |
| This function calculates the GETAWAY descriptors for a molecule, which capture the shape and size of the molecule. | Computational Tools | |
| This function calculates the Hall-Kier alpha index for a molecule, which is a measure of its shape. | Computational Tools | |
| This function calculates the Inertial Shape Factor of a molecule, which is a measure of its shape. | Computational Tools | |
| This function computes the Kappa1 ($`K1`$) value of a molecule, which is a topological descriptor representing its shape complexity or branching degree. | Computational Tools | |
| This function computes the Kappa2 ($`K2`$) value of a molecule, which is a topological descriptor representing its shape complexity or branching degree. | Computational Tools | |
| This function computes the Kappa3 ($`K3`$) value of a molecule, which is a topological descriptor representing its shape complexity or branching degree. | Computational Tools | |
| This function calculates the Labute accessible surface area (ASA) value for a molecule, which is a measure of the solvent-accessible surface area of the molecule. | Computational Tools | |
| This function calculates the molecular formula of a molecule, which is a string representing the number and type of atoms in the molecule. | Computational Tools | |
| This tool calculates the Molecule Representation of Structures based on Electron diffraction (MORSE) descriptors for a given molecule. | Computational Tools | |
| This function calculates the NPR1 (Normalized Principal Moments Ratio) descriptor for a molecule, which serves as a descriptor for the distribution of charges within the molecule. | Computational Tools | |
| This function calculates the NPR2 (Normalized Principal Moments Ratio) descriptor for a molecule, which serves as a descriptor for the distribution of charges within the molecule. | Computational Tools | |
| This function calculates the number of aliphatic carbocycles in a molecule. | Computational Tools | |
| This function calculates the number of aliphatic heterocycles in a molecule. | Computational Tools | |
| This tool calculates the number of aliphatic rings in a molecule. | Computational Tools | |
| This function calculates the number of amide bonds in a molecule. | Computational Tools | |
| This function calculates the number of aromatic carbocycles in a molecule. | Computational Tools | |
| This function calculates the number of aromatic heterocycles in a molecule. | Computational Tools | |
| This tool calculates the number of aromatic rings in a molecule. | Computational Tools | |
| This function calculates the number of atom stereo centers in a molecule. | Computational Tools | |
| This function calculates the number of atoms in a molecule. | Computational Tools | |
| This function calculates the number of bridgehead atoms in a molecule. | Computational Tools | |
| This function calculates the number of hydrogen bond acceptors (HBA) in a molecule. | Computational Tools | |
| This function calculates the number of hydrogen bond donors (HBD) in a molecule. | Computational Tools | |
| This tool calculates the number of heavy atoms in a molecule. | Computational Tools | |
| This tool calculates the number of heteroatoms in a molecule. | Computational Tools | |
| This tool calculates the number of heterocycles in a molecule. | Computational Tools | |
| This tool calculates the number of Lipinski hydrogen bond acceptors (HBA) in a molecule, which is a measure used in drug-likeness evaluation according to Lipinski’s rule of five. | Computational Tools | |
| This tool calculates the number of Lipinski hydrogen bond donors (HBD) in a molecule, which is a measure used in drug-likeness evaluation according to Lipinski’s rule of five. | Computational Tools | |
| This tool calculates the number of rings in a molecule. | Computational Tools | |
| This tool calculates the number of rotatable bonds in a molecule. | Computational Tools | |
| This function calculates the number of saturated carbocycles in a molecule. | Computational Tools | |
| This function calculates the number of saturated heterocycles in a molecule. | Computational Tools | |
| This tool calculates the number of saturated rings in a molecule. | Computational Tools | |
| This function calculates the number of spiro atoms in a molecule. | Computational Tools | |
| This tool calculates the number of unspecified atomic stereocenters in a molecule. | Computational Tools | |
| Generate RDKfingerprints for the SMILES strings in a CSV file and save to a new CSV file. | Computational Tools | |
| Generate morgan fingerprints for the SMILES strings in a CSV file and save to a new CSV file. | Computational Tools | |
| Generate electrical RDKit descriptors for the SMILES strings in a CSV file and save to a new CSV file. | Computational Tools | |
| General MLP classifier function that predicts based on processed feature files. | Computational Tools | |
| General AdaBoost classifier function that predicts based on processed feature files. | Computational Tools | |
| General Random Forest classifier function that predicts based on processed feature files. | Computational Tools | |
| Adds the oxidation number/state to the atoms of a molecule as property OxidationNumber on each atom. | Computational Tools | |
| This tool calculates the PBF (plane of best fit) descriptor for a given molecule. | Computational Tools | |
| This tool calculates the first principal moment of inertia (PMI1) for a given molecule. | Computational Tools | |
| This tool is designed to compute the PMI2 (Partial Molecular Information 2) value of a molecule, which serves as a descriptor indicating the shape and structure of the molecule. | Computational Tools | |
| This tool is designed to compute the PMI3 (Partial Molecular Information 3) value of a molecule, which serves as a descriptor characterizing the shape and structure of the molecule. | Computational Tools | |
| This tool calculates the Phi ($`\Phi`$) angle of a molecule, which is a torsional angle describing the rotation about a single bond. | Computational Tools | |
| This tool calculates the RDF (Radial Distribution Function) descriptor for a given molecule. | Computational Tools | |
| This tool is designed to compute the radius of gyration for a given molecule, providing insights into its overall shape and compactness. | Computational Tools | |
| This function calculates the sphericity index for a given molecule. | Computational Tools | |
| This tool calculates the TPSA (Topological Polar Surface Area) descriptor for a given molecule, which is a measure of the accessible polar surface area in a molecule. | Computational Tools | |
| This tool calculates the WHIM (Weighted Holistic Invariant Molecular) descriptor for a given molecule. | Computational Tools | |
| This function computes a custom property for a given molecule using the Van der Waals Surface Area (VSA) method, based on user-defined parameters. | Computational Tools | |
| This function computes a set of atom features for a given molecule, including atomic number, valence, and hybridization. | Computational Tools | |
| This function computes the atom pair for a given molecule. | Computational Tools | |
| This tool computes connectivity invariants, similar to ECFP (Extended Connectivity Fingerprints), for a given molecule. | Computational Tools | |
| This tool computes feature invariants, similar to FCFP (Feature Centroid Fingerprints), for a given molecule. | Computational Tools | |
| This function computes atom pair code (hash) for each atom in a molecular. | Computational Tools | |
| This function computes the hybridization of each atom in a molecule. | Computational Tools | |
| This function computes the ring systems of a molecule. | Computational Tools | |
| This function computes the Molecular ACCess System keys fingerprint for a given molecule. | Computational Tools | |
| This tool computes the Morgan fingerprint for a given molecule. | Computational Tools | |
| This tool computes the topological torsion fingerprint for a given molecule. | Computational Tools | |
| The tool computes the USR (Ultrafast Shape Recognition) descriptor for a given conformer of a molecule and returns it as a list. | Computational Tools | |
| This function is designed to compute the USRCAT (Ultrafast Shape Recognition with Coordinate Asymmetric Torsions) descriptor for a specified conformer of a molecule. | Computational Tools | |
| This function is used to add hydrogen atoms to the molecular graph of a molecule. | Computational Tools | |
| This tool adds wavy bonds around double bonds with STEREOANY stereochemistry. | Computational Tools | |
| This tool sets chiral tags for atoms of the molecular based on the molParity property. | Computational Tools | |
| This tool is used to assign radical counts to atoms within a molecule. | Computational Tools | |
| This tool is used for assigning Cahn–Ingold–Prelog (CIP) stereochemistry to atoms (R/S) and double bonds (Z/E) within a molecule. | Computational Tools | |
| This tool is used to obtain the adjacency matrix of a molecule. | Computational Tools | |
| This tool is used to determine whether recognition of non-tetrahedral chirality from 3D structures is enabled or not. | Computational Tools | |
| The tool computes the topological distance matrix for a given molecule. | Computational Tools | |
| This tool is utilized to determine the total formal charge of a given molecule. | Computational Tools | |
| This tool is utilized to determine the formal charge of each atom in a given molecule. | Computational Tools | |
| This tool identifies disconnected fragments within a molecule and returns them as atom identifiers or molecules. | Computational Tools | |
| This tool is used to determine whether the legacy stereo perception code is being used. | Computational Tools | |
| This tool is used to convert a molecule that represents haptic bonds using a dummy atom with a dative bond to a metal atom into a molecule with explicit dative bonds from the atoms of the haptic group to the metal atom. | Computational Tools | |
| This tool is used to check if a molecule contains query H (hydrogen) atoms. | Computational Tools | |
| This tool is used to perform Kekulization on a molecule. | Computational Tools | |
| This tool is used to merge hydrogen atoms into their neighboring atoms as query atoms. | Computational Tools | |
| This tool is used to perform a Murcko decomposition on a molecule and return the scaffold. | Computational Tools | |
| This tool is used to remove hydrogen atoms from a molecule’s graph. | Computational Tools | |
| This tool is used to remove all stereochemistry information from a molecule. | Computational Tools | |
| This tool is used to perform aromaticity perception on a molecule, which means determining the aromaticity of atoms and bonds in the molecule. | Computational Tools | |
| This tool is used to set the cis/trans stereochemistry on double bonds based on the directions of neighboring bonds. | Computational Tools | |
| This tool is used to check if a molecule(target) is a substructure of another molecule(template). | Computational Tools | |
| Generate pre-signed URLs for uploading and downloading Alibaba Cloud OSS objects | Computational Tools | |
| Query UniProt for protein information. | Databases | |
| Query InterPro for protein domain information. | Databases | |
| Download PDB structure file. | Databases | |
| Download NCBI sequence file. | Databases | |
| Download AlphaFold structure file. | Databases | |
| Retrieve and download the protein sequence (. | Databases | |
| Retrieve SMILES strings from PubChem using compound names. | Databases | |
| Search UniProt ID by gene name. | Databases | |
| Download predicted protein structures from the AlphaFold database. | Databases | |
| Retrieve the details of a single bioactivity entry from the ChEMBL database by its unique activity ID. | Databases | |
| Retrieves a list of bioactivity entries from the ChEMBL database. | Databases | |
| Performs a full-text search for bioactivity data in the ChEMBL database using a query string. | Databases | |
| Retrieves a default list of supplementary bioactivity data from the ChEMBL database. | Databases | |
| Retrieve single activitysupplementarydatabyactivity object details by ID. | Databases | |
| Retrieve multiple activitysupplementarydatabyactivity objects by IDs. | Databases | |
| Retrieves the details for a single assay (experimental procedure) from the ChEMBL database using its name. | Databases | |
| Retrieves detailed information for multiple assays from the ChEMBL database using a list of their names. | Databases | |
| Performs a full-text search for assays (experimental procedures) in the ChEMBL database using a query string. | Databases | |
| Retrieves a default list of assay classifications from the ChEMBL database. | Databases | |
| Retrieves the details for a single ATC (Anatomical Therapeutic Chemical) classification from the ChEMBL database using the level 5 ATC code. | Databases | |
| Retrieves the details for multiple ATC (Anatomical Therapeutic Chemical) classifications from the ChEMBL database using a list of their level 5 ATC codes. | Databases | |
| Retrieves the details for a single binding site from the ChEMBL database using its unique integer ID. | Databases | |
| Retrieves detailed information for multiple binding sites from the ChEMBL database using a list of their unique integer IDs. | Databases | |
| Retrieves the details for a single biotherapeutic from the ChEMBL database using its name. | Databases | |
| Retrieves detailed information for multiple biotherapeutics from the ChEMBL database using a list of their names. | Databases | |
| Retrieves the details for a single cell line from the ChEMBL database using its unique integer ID. | Databases | |
| Retrieves detailed information for multiple cell lines from the ChEMBL database using a list of their unique integer IDs. | Databases | |
| Retrieve compound structural alert object list. | Databases | |
| Retrieve compound structural alert object details by ID. | Databases | |
| Retrieve multiple compound structural alert objects by IDs. | Databases | |
| Retrieve single drug object details by name. | Databases | |
| Retrieve multiple drug objects by names. | Databases | |
| Retrieve drug indication object details by ID | Databases | |
| Retrieve multiple drug indication objects by IDs. | Databases | |
| Retrieve single drug_warning object details by ID | Databases | |
| Retrieve multiple drug_warning objects by IDs. | Databases | |
| Retrieves the details for a single GO (Gene Ontology) slim classification from the ChEMBL database using its unique GO ID. | Databases | |
| Retrieves the details for multiple GO (Gene Ontology) slim classifications from the ChEMBL database using a list of their unique GO IDs. | Databases | |
| Retrieves the details for a single drug mechanism of action from the ChEMBL database using its unique integer ID. | Databases | |
| Retrieves detailed information for multiple drug mechanisms of action from the ChEMBL database using a list of their unique integer IDs. | Databases | |
| Retrieve single metabolism object details by ID. | Databases | |
| Retrieves detailed information for multiple drug metabolism records from the ChEMBL database using a list of their unique integer IDs. | Databases | |
| Retrieves the details for a single molecule from the ChEMBL database using its name. | Databases | |
| Retrieves detailed information for multiple molecules from the ChEMBL database using a list of their names. | Databases | |
| Retrieves a default list of molecule forms from the ChEMBL database. | Databases | |
| Retrieves molecule form information for a given molecule from the ChEMBL database using its name. | Databases | |
| Retrieves molecule form information for multiple molecules from the ChEMBL database using a list of their names. | Databases | |
| Retrieves the details for a single organism from the ChEMBL database using its unique integer ID. | Databases | |
| Retrieves detailed information for multiple organisms from the ChEMBL database using a list of their unique integer IDs. | Databases | |
| Retrieves the details for a single protein classification from the ChEMBL database using its unique integer ID. | Databases | |
| Retrieves detailed information for multiple protein classifications from the ChEMBL database using a list of their unique integer IDs. | Databases | |
| Search protein_classification object by query string. | Databases | |
| Retrieve similarity details for compounds based on SMILES. | Databases | |
| Retrieve single source object details by ID. | Databases | |
| Retrieve multiple source object details by IDs. | Databases | |
| Retrieve substructure matches using SMILES. | Databases | |
| Retrieve single target object details by name. | Databases | |
| Retrieve multiple target objects by names | Databases | |
| Search target using query string. | Databases | |
| Retrieve target_component object list | Databases | |
| Retrieve single target_relation object details by related target name | Databases | |
| Retrieve multiple target_relation objects by related target names | Databases | |
| Retrieve single tissue object details by ID. | Databases | |
| Retrieve multiple tissue object details by IDs. | Databases | |
| Get compound chembl id by name. | Databases | |
| Get target chembl id by name. | Databases | |
| Get assay chembl id by name. | Databases | |
| This operation displays the database release information with statistics for the databases. | Databases | |
| KEGG find - Data search. | Databases | |
| This operation can be used to obtain a list of all entries in each database. | Databases | |
| This operation retrieves given database entries in a flat file format or in other formats with options. | Databases | |
| This operation can be used to convert entry identifiers (accession numbers) of outside databases to KEGG identifiers, and vice versa. | Databases | |
| KEGG link - find related entries by using database cross-references. | Databases | |
| Maps common protein names, synonyms and UniProt identifiers into STRING identifiers | Databases | |
| Retrieve STRING interaction network for one or multiple proteins in various text formats. | Databases | |
| This method provides the interactions between your provided set of proteins and all the other STRING proteins. | Databases | |
| STRING internally uses the Smith-Waterman bit scores as a proxy for protein homology. | Databases | |
| Retrieve the similarity from your input protein(s) to the best (most) similar protein from each STRING species. | Databases | |
| STRING maps several databases onto its proteins, this includes: Gene Ontology, KEGG pathways, UniProt Keywords, PubMed publications, Pfam domains, InterPro domains, and SMART domains. | Databases | |
| STRING maps several databases onto its proteins, this includes: Gene Ontology, KEGG pathways, UniProt Keywords, PubMed publications, Pfam domains, InterPro domains, and SMART domains. | Databases | |
| Get protein-protein interaction enrichment for list of genes denoted by their STRING identifiers | Databases | |
| Providing Information of variants of human proteins, and whether these variants are benign, mutual or harmful. | Databases | |
| Database containing gene, gene set and collection of gene sets information. | Databases | |
| A database containing the relationship between mouse gene sets and diseases. | Databases | |
| Ensemble is a database containing the relationship between human genes and chr locations. | Databases | |
| Search PubChem for compounds matching a chemical name. | Databases | |
| Search PubChem for compounds matching a SMILES string. | Databases | |
| Get detailed compound information by PubChem CID. | Databases | |
| Perform an advanced search on PubChem using a complex query. | Databases | |
| Get substance information by PubChem SID. | Databases | |
| Get compound information by PubChem CID. | Databases | |
| Get compound information by chemical name. | Databases | |
| Get substance information by name. | Databases | |
| Retrieve a specific chemical property for a compound by its name from PubChem. | Databases | |
| Retrieve all known synonyms for a given compound by its chemical name from PubChem. | Databases | |
| Get detailed description information for a PubChem substance given its SID. | Databases | |
| Retrieve detailed description information for a PubChem compound given its CID. | Databases | |
| Get detailed description of a compound by name, including overall information, drug and medication information, pharmacology and biochemistry information. | Databases | |
| Retrieve detailed description information for a PubChem bioassay given its AID. | Databases | |
| Retrieve a summary of bioassay activities for a given PubChem compound CID. | Databases | |
| Retrieve a summary of bioassay activities for a given PubChem substance SID. | Databases | |
| Get summary information for a gene by Gene ID. | Databases | |
| Retrieve summary information for a protein from PubChem by its UniProt or other accession number. | Databases | |
| Retrieve summary information for a biological taxonomy entry given its NCBI Taxonomy ID. | Databases | |
| Retrieve available conformer identifiers for a given PubChem compound CID. | Databases | |
| Retrieve compound objects from PubChem based on a given SMILES string. | Databases | |
| Retrieve compound objects from PubChem based on a molecular formula. | Databases | |
| Get the molecular formula of a compound. | Databases | |
| Get the molecular weight of a compound. | Databases | |
| Get the isomeric SMILES of a compound. | Databases | |
| Get the XLogP value of a compound. | Databases | |
| Get the IUPAC name of a compound. | Databases | |
| Get the synonyms of a compound. | Databases | |
| Obtain the CID corresponding to the drug smiles | Databases | |
| Get a list of CIDs by molecular formula. | Databases | |
| Get a list of SIDs by name. | Databases | |
| Get a Substance object by SID using PubChemPy. | Databases | |
| Get a list of Substance objects by name using PubChemPy. | Databases | |
| Get the source ID (Unique identifier assigned to the compound or substance by the original database (e. | Databases | |
| Get the synonyms (Different names or identifiers for the same chemical substance) of a substance by SID. | Databases | |
| Get a dictionary of a compound’s properties. | Databases | |
| Get a list of compound objects with 3D structures. | Databases | |
| Get a dictionary representation of a compound by CID. | Databases | |
| Get CAS Registry Numbers for compounds containing a specified substructure. | Databases | |
| Get a gene summary by gene symbol. | Databases | |
| Get gene information by gene IDs. | Databases | |
| Get gene information by accession. | Databases | |
| Get dataset reports by accession IDs | Databases | |
| Get gene product reports by accession IDs | Databases | |
| Get gene download summary by GeneID | Databases | |
| Get gene links by gene ID | Databases | |
| Get gene dataset reports by locus tag | Databases | |
| Get gene product reports by locus tags | Databases | |
| Get dataset reports by taxons | Databases | |
| Get product reports by taxon | Databases | |
| Get gene dataset reports by taxonomic identifier | Databases | |
| Get gene product reports by taxonomic identifier | Databases | |
| Get gene information by dataset report | Databases | |
| Get genome annotation reports by genome accession. | Databases | |
| Get genome annotation report summary information. | Databases | |
| Get revision history for assembly by accession | Databases | |
| Get sequence reports by accessions | Databases | |
| Check the validity of genome accessions | Databases | |
| Get dataset reports by accessions | Databases | |
| Get a genome dataset by accession | Databases | |
| Preview genome dataset download | Databases | |
| Get assembly links by accessions | Databases | |
| Get dataset reports by assembly name | Databases | |
| Get dataset reports by bioproject | Databases | |
| Get dataset reports by biosample id | Databases | |
| Get assembly accessions for a sequence accession | Databases | |
| Get dataset reports by taxons | Databases | |
| Get dataset reports by wgs accession | Databases | |
| Get virus annotation report by accessions. | Databases | |
| Check virus accessions validity | Databases | |
| Get virus dataset report by accessions | Databases | |
| Get virus annotation report by taxon | Databases | |
| Get virus genome by taxon | Databases | |
| Get virus genome table by taxon | Databases | |
| Get related taxonomy IDs | Databases | |
| Get taxonomy links | Databases | |
| Get taxonomy information. | Databases | |
| Get taxonomy dataset report. | Databases | |
| Get filtered taxonomy subtree | Databases | |
| Get taxonomy name report | Databases | |
| Get taxonomy suggestions | Databases | |
| Get biosample report. | Databases | |
| Get organelle dataset report. | Databases | |
| Get organelle dataset report by taxon. | Databases | |
| Get gene product report by gene ID | Databases | |
| Get gene orthologs by gene ID | Databases | |
| Get gene information by taxon | Databases | |
| Get gene counts by taxon | Databases | |
| Get chromosome summary by taxon and annotation name | Databases | |
| Get genome information by accession | Databases | |
| Get a prokaryote gene dataset by RefSeq protein accession | Databases | |
| Get general information of a protein or gene by name from UniProt database. | Databases | |
| Get the human protein sequence by protein name. | Databases | |
| Search UniProtKB by protein entry accession to return all data associated with that entry. | Databases | |
| Stream all UniProtKB entries associated with the search term in a single download. | Databases | |
| Search UniProtKB entries using a query, returns paginated list. | Databases | |
| Search UniRef entry by id to return all data associated with that entry. | Databases | |
| Search UniRef entry by member id to return all data associated with that entry. | Databases | |
| Search light UniRef entry by id to return all data associated with that entry. | Databases | |
| Stream all UniRef clusters associated with the search term in a single download. | Databases | |
| Search UniRef clusters using a query, returns paginated list. | Databases | |
| Search UniParc entry by id (UPI) to return all data associated with that entry. | Databases | |
| Search UniParc entry by id (UPI) to return all data associated with that entry (light version). | Databases | |
| Get a page of database cross-reference entries by a UPI. | Databases | |
| Stream database cross-reference entries for a specified UniParc UPI. | Databases | |
| Stream all UniParc entries associated with the specified search term in a single download. | Databases | |
| Search UniParc entries using a query, returns paginated list. | Databases | |
| Retrieve a GeneCentric entry by UniProtKB accession. | Databases | |
| Search GeneCentric entry by Proteome ID to return all data associated with that entry. | Databases | |
| Stream GeneCentric entries matching a query (max 10M entries). | Databases | |
| Search GeneCentric entries with pagination. | Databases | |
| Retrieve a proteome by UniProt Proteome ID. | Databases | |
| Stream Proteome entries matching a query (max 10M entries). | Databases | |
| Search Proteome entries with pagination. | Databases | |
| Analyze the tissue-specific expression pattern of a given gene across different cancer types using the Firebrowse API (TCGA mRNASeq). | Databases | |
| Look up Ensembl gene information by external gene symbol. | Databases | |
| Find evolutionary homologs (orthologs and paralogs) for a gene identified by symbol. | Databases | |
| Retrieve genomic DNA sequence for a specific chromosomal region. | Databases | |
| Predict the functional effects of variants using Variant Effect Predictor (VEP) with HGVS notation. | Databases | |
| Retrieve a phylogenetic gene tree by its Ensembl stable identifier. | Databases | |
| Retrieve genome assembly information for a species. | Databases | |
| Get cross references for a gene symbol. | Databases | |
| Get the latest version of an Ensembl stable identifier. | Databases | |
| Get the latest version for multiple Ensembl stable identifiers. | Databases | |
| Retrieve a CAFE (Computational Analysis of gene Family Evolution) gene tree by ID. | Databases | |
| Retrieve a CAFE gene tree for a gene identified by symbol. | Databases | |
| Retrieve the gene tree containing a gene identified by its Ensembl ID. | Databases | |
| Get gene tree by symbol. | Databases | |
| Retrieve genomic alignments between species for a specific region. | Databases | |
| Find evolutionary homologs (orthologs and paralogs) for a gene identified by Ensembl ID. | Databases | |
| Get cross references by ID. | Databases | |
| Get cross references by name. | Databases | |
| List the analyses and data processing pipelines used for a species genome. | Databases | |
| Retrieve detailed information about a specific genomic region or chromosome. | Databases | |
| Retrieve the catalog of gene and transcript biotypes for a species. | Databases | |
| Get external databases for a species. | Databases | |
| Map cDNA coordinates to genomic coordinates. | Databases | |
| Map protein coordinates to genomic coordinates. | Databases | |
| Get ontology ancestors. | Databases | |
| Get ontology descendants. | Databases | |
| Get ontology by ID. | Databases | |
| Get ontology by name. | Databases | |
| Get features overlapping a region defined by an identifier. | Databases | |
| Get features overlapping a genomic region. | Databases | |
| Get features overlapping a translation. | Databases | |
| Retrieve phenotype associations for variants in a genomic region. | Databases | |
| Retrieve phenotype associations for a specific gene. | Databases | |
| Retrieve genomic features associated with a specific phenotype ontology term. | Databases | |
| Retrieve sequence associated with an Ensembl identifier. | Databases | |
| Predict the functional effects of variants using Variant Effect Predictor (VEP) with variant identifier. | Databases | |
| Predict the functional effects of variants using Variant Effect Predictor (VEP) with genomic coordinates. | Databases | |
| Retrieve detailed information about a genetic variant by its identifier. | Databases | |
| Find information about a given genome. | Databases | |
| Find information about genomes containing a specified INSDC accession. | Databases | |
| Find information about a genome with a specified assembly. | Databases | |
| Find information about all genomes in a given division. | Databases | |
| Find information about all genomes beneath a given node of the taxonomy. | Databases | |
| List all variation data sources used for a species in Ensembl. | Databases | |
| Lists all variant consequence types used by Ensembl. | Databases | |
| List all variation populations for a species, or list all individuals in a specific population. | Databases | |
| Look up details for any Ensembl stable identifier. | Databases | |
| Look up details for multiple Ensembl stable identifiers in a single request. | Databases | |
| Look up multiple gene symbols in a single request. | Databases | |
| Reconstruct the entire ancestry of a term from is_a and part_of relationships. | Databases | |
| Return the taxonomic classification of a taxon node. | Databases | |
| Search for a taxonomic term by its identifier or name | Databases | |
| Search for a taxonomic id by a non-scientific name. | Databases | |
| Return the specified binding matrix | Databases | |
| Request multiple types of sequence by a stable identifier list. | Databases | |
| Get sequences by multiple regions. | Databases | |
| Batch predict the functional effects of multiple variants using VEP with variant identifiers. | Databases | |
| Get variant effect predictions by multiple regions. | Databases | |
| Translate a list of variant identifiers, HGVS notations or genomic SPDI notations to all possible variant IDs, HGVS and genomic SPDI | Databases | |
| Fetch variants by publication using PubMed Central reference number (PMCID) | Databases | |
| Fetch variants by publication using PubMed reference number (PMID) | Databases | |
| Uses a list of variant identifiers (e. | Databases | |
| Get Beacon information. | Databases | |
| Query Beacon. | Databases | |
| Query Beacon with POST. | Databases | |
| Get GA4GH features by ID. | Databases | |
| Get a list of sequence annotation features in GA4GH format | Databases | |
| Search GA4GH callsets. | Databases | |
| Get the GA4GH record for a specific CallSet given its identifier | Databases | |
| Get a list of datasets in GA4GH format | Databases | |
| Get the GA4GH record for a specific dataset given its identifier | Databases | |
| Search GA4GH feature sets. | Databases | |
| Return the GA4GH record for a specific featureSet given its identifier | Databases | |
| Get GA4GH variant by ID. | Databases | |
| Return variant annotation information in GA4GH format for a region on a reference sequence | Databases | |
| Return variant call information in GA4GH format for a region on a reference sequence | Databases | |
| Search GA4GH variant sets. | Databases | |
| Return the GA4GH record for a specific VariantSet given its identifier | Databases | |
| Return a list of reference sequences in GA4GH format | Databases | |
| Return data for a specific reference in GA4GH format by id | Databases | |
| Search GA4GH reference sets. | Databases | |
| Search data for a specific reference set in GA4GH format by ID | Databases | |
| Return a list of annotation sets in GA4GH format | Databases | |
| Return meta data for a specific annotation set in GA4GH format by ID | Databases | |
| Retrieve the gene tree containing a gene identified by its Ensembl ID. | Databases | |
| With :group argument provided, list the properties of biotypes within that group. | Databases | |
| List the properties of biotypes with a given name. | Databases | |
| List all collections of species analysed with the specified compara method. | Databases | |
| Get all available comparative genomics databases and their data release. | Databases | |
| Get all supported genome assemblies from UCSC Genome Browser. | Databases | |
| List all tracks for a specific genome assembly. | Databases | |
| List all tracks in a specific track hub for a genome. | Databases | |
| List all chromosomes for a genome assembly. | Databases | |
| Get list of all public UCSC track hubs. | Databases | |
| Get sequence for an entire chromosome. | Databases | |
| Get DNA sequence for a genomic region. | Databases | |
| Get cytoband (chromosome banding) information for a specified genome and chromosome. | Databases | |
| Fetch a list of active ingredients in a specific drug product. | Databases | |
| Retrieve dosage and storage information for a specific drug. | Databases | |
| Retrieve drug names based on information about types of abuse and adverse reactions pertinent to those types of abuse. | Databases | |
| Retrieve information about types of abuse based on the drug name. | Databases | |
| Retrieve drug names based on the accessories field information. | Databases | |
| Retrieve information about accessories based on the drug name. | Databases | |
| Retrieve drug names based on the active ingredient information. | Databases | |
| Retrieve detailed information about a drug’s active ingredient, FDA application number, manufacturer name, National Drug Code (NDC) number, and route of administration; all based on the drug name. | Databases | |
| Retrieve drug names based on the specified FDA application number, manufacturer name, or National Drug Code (NDC) number. | Databases | |
| Retrieve the drug name based on specific adverse reactions reported. | Databases | |
| Retrieve adverse reactions information based on the drug name. | Databases | |
| Retrieve drug names based on the presence of specific alarms, which are related to adverse reaction events. | Databases | |
| Retrieve alarms based on the specified drug name. | Databases | |
| Retrieve drug names based on animal pharmacology and toxicology information. | Databases | |
| Retrieve animal pharmacology and toxicology information based on drug names. | Databases | |
| Retrieve the drug names that require asking a doctor before use due to a patient’s specific conditions and symptoms. | Databases | |
| Get information about when a doctor should be consulted before using a specific drug. | Databases | |
| Retrieve drug names based on information about when a doctor or pharmacist should be consulted regarding drug interactions. | Databases | |
| Get information about when a doctor or pharmacist should be consulted regarding drug interactions for a specific drug. | Databases | |
| Retrieve drug names based on assembly or installation instructions. | Databases | |
| Retrieve assembly or installation instructions based on drug names. | Databases | |
| Retrieve drug names that have specific boxed warnings (The most serious risk alerts in drug labeling) and adverse effects. | Databases | |
| Retrieve boxed warning (The most serious risk alerts in drug labeling) and adverse effects information for a specific drug. | Databases | |
| Retrieve the drug name based on the calibration instructions provided. | Databases | |
| Retrieve calibration instructions based on the specified drug name. | Databases | |
| Retrieve drug names based on the presence of carcinogenic, mutagenic, or fertility impairment information. | Databases | |
| Retrieve carcinogenic, mutagenic, or fertility impairment information based on the drug name. | Databases | |
| Retrieves the drug name based on various identifiers such as the FDA application number, NUI, SPL document ID, or SPL set ID. | Databases | |
| Retrieves drug names based on a search query within their clinical pharmacology information. | Databases | |
| Retrieves clinical pharmacology information for a specific drug from the FDA database by its name. | Databases | |
| Retrieves drug names based on a search query within their clinical studies information. | Databases | |
| Retrieves clinical studies information for a specific drug from the FDA database by its name. | Databases | |
| Retrieves drug names based on a search query within their contraindications information. | Databases | |
| Retrieve contraindications information based on the drug name. | Databases | |
| Retrieves drug names based on a search query within their DEA controlled substance schedule information. | Databases | |
| Retrieves information about the controlled substance DEA (Drug Enforcement Administration) schedule for a specific drug. | Databases | |
| Retrieve the drug name based on information about dependence characteristics. | Databases | |
| Retrieves information about dependence characteristics for a specific drug from the FDA database by its name. | Databases | |
| Retrieves drug names based on a search query within their disposal and waste handling information. | Databases | |
| Retrieves disposal and waste handling information for a specific drug from the FDA database by its name. | Databases | |
| Retrieve the drug name based on dosage and administration information. | Databases | |
| Retrieves drug names based on a search query within their dosage forms and strengths information. | Databases | |
| Retrieves dosage forms and strengths information for a specific drug from the FDA database by its name. | Databases | |
| Retrieves the drug name based on information about drug abuse and dependence, including whether the drug is a controlled substance, the types of possible abuse, and adverse reactions relevant to those abuse types. | Databases | |
| Retrieves information about drug abuse and dependence for a specific drug from the FDA database by its name, including controlled substance status, abuse types, and related adverse reactions. | Databases | |
| Retrieves drug names based on a search query within their laboratory test interference information. | Databases | |
| Retrieves information about laboratory test interferences for a specific drug. | Databases | |
| Retrieves a list of drug names that have known interactions with a specified term. | Databases | |
| Retrieve drug interactions based on the specified drug name. | Databases | |
| Retrieve drug names based on the effective time of the labeling document. | Databases | |
| Retrieve effective time of the labeling document based on the drug name. | Databases | |
| Retrieve the drug name based on the specified environmental warnings. | Databases | |
| Fetch environmental warnings for a specific drug based on its name. | Databases | |
| Retrieve drug names based on specific food safety warnings. | Databases | |
| Retrieve drug names based on specific general precautions information. | Databases | |
| Retrieve general precautions information based on the drug name. | Databases | |
| Retrieve drug names that have specific information about geriatric use. | Databases | |
| Retrieve information about geriatric use based on the drug name. | Databases | |
| Fetch information about dear health care provider letters for a specific drug. | Databases | |
| Fetch drug names based on information about dear health care provider letters. | Databases | |
| Retrieve drug names based on specific health claims. | Databases | |
| Retrieve health claims associated with a specific drug name. | Databases | |
| Retrieve the drug name based on the document ID. | Databases | |
| Retrieve the document ID based on the drug name. | Databases | |
| Retrieve the drug name based on the inactive ingredient information. | Databases | |
| Fetch a list of inactive ingredients in a specific drug product based on the drug name. | Databases | |
| Retrieve a list of drug names based on a specific indication or usage. | Databases | |
| Retrieve indications and usage information based on a specific drug name. | Databases | |
| Retrieve drug names based on information for owners or caregivers. | Databases | |
| Retrieves information for owners or caregivers for a specific drug. | Databases | |
| Retrieves patient information for a specific drug from the FDA database by its name. | Databases | |
| Retrieves drug names based on a search query within their instructions for use. | Databases | |
| Retrieves instructions for use information for a specific drug from the FDA database by its name. | Databases | |
| Retrieves drug names based on a search query within the intended use of their associated medical device. | Databases | |
| Retrieves the intended use of the device associated with a specific drug. | Databases | |
| Retrieves drug names based on a search query within their child safety information. | Databases | |
| Retrieves child safety information for a specific drug based on its name. | Databases | |
| Retrieve the drug name based on information about the drug’s use during labor or delivery. | Databases | |
| Retrieves information about the drug’s use during labor or delivery based on the drug name. | Databases | |
| Retrieves drug names based on a search query within their laboratory tests information. | Databases | |
| Retrieves laboratory tests information for a specific drug from the FDA database by its name. | Databases | |
| Retrieves the mechanism of action information for a specific drug. | Databases | |
| Retrieves drug names based on a search query within their mechanism of action information. | Databases | |
| Retrieve the drug name based on microbiology field information. | Databases | |
| Retrieves microbiology information for a specific drug from the FDA database by its name. | Databases | |
| Retrieves drug names based on a search query within their nonclinical toxicology information. | Databases | |
| Retrieves nonclinical toxicology information for a specific drug from the FDA database by its name. | Databases | |
| Retrieve drug names based on the presence of nonteratogenic effects information. | Databases | |
| Retrieves information about nonteratogenic effects for a specific drug from the FDA database by its name. | Databases | |
| Retrieves drug names based on a search query within their information for nursing mothers. | Databases | |
| Retrieves information about nursing mothers for a specific drug. | Databases | |
| Retrieves drug names based on a search query within their ’other safety information’ section. | Databases | |
| Retrieves other safety information for a specific drug from the FDA database by its name. | Databases | |
| Retrieves drug names based on a search query within their overdosage information. | Databases | |
| Retrieves information about signs, symptoms, and laboratory findings of acute overdosage based on the drug name. | Databases | |
| Retrieve the drug name based on the content of the principal display panel of the product package. | Databases | |
| Retrieve the content of the principal display panel of the product package based on the drug name. | Databases | |
| Retrieve drug names based on patient medication information, which is about safe use of the drug. | Databases | |
| Retrieve patient medication information (which is about safe use of the drug) based on drug names. | Databases | |
| Retrieve drug names based on pediatric use information. | Databases | |
| Retrieve pediatric use information based on drug names. | Databases | |
| Retrieve the drug name based on pharmacodynamics information. | Databases | |
| Retrieve pharmacodynamics information based on the drug name. | Databases | |
| Retrieve pharmacogenomics information based on the drug name. | Databases | |
| Retrieve drug names based on specific pharmacokinetics information, such as absorption, distribution, elimination, metabolism, drug interactions, and specific patient populations. | Databases | |
| Retrieve pharmacokinetics information (e. | Databases | |
| Retrieve the drug name based on the precautions field information. | Databases | |
| Retrieve precautions information based on the drug name. | Databases | |
| Retrieves drug names based on a search query within their pregnancy effects information. | Databases | |
| Retrieves information about the effects on pregnancy for a specific drug. | Databases | |
| Retrieve the drug names based on pregnancy or breastfeeding information. | Databases | |
| Retrieves pregnancy or breastfeeding information for a specific drug. | Databases | |
| Retrieves information on who to contact with questions about the drug based on the provided drug name. | Databases | |
| Retrieves recent major changes in labeling for a specific drug. | Databases | |
| Retrieves the drug name based on the reference information provided in the drug labeling. | Databases | |
| Retrieves reference information for a specific drug from the FDA database by its name. | Databases | |
| Retrieves drug names based on a search query within their residue warning information. | Databases | |
| Retrieves the residue warning for a specific drug from the FDA database by its name. | Databases | |
| Retrieves drug names based on a search query within their risk information, especially regarding pregnancy or breastfeeding. | Databases | |
| Retrieves risk information (especially regarding pregnancy or breastfeeding) for a specific drug from the FDA database by its name. | Databases | |
| Retrieves drug names based on a search query within their route of administration. | Databases | |
| Retrieve the route of administration information based on the drug name. | Databases | |
| Retrieves drug names based on a search query within their safe handling warning information. | Databases | |
| Retrieves safe handling warnings for a specific drug from the FDA database by its name. | Databases | |
| Retrieve the drug name based on the Set ID of the labeling. | Databases | |
| Retrieve drug names based on Structured Product Labeling (SPL) indexing data elements. | Databases | |
| Retrieve Structured Product Labeling (SPL) indexing data elements based on drug names. | Databases | |
| Retrieve drug names based on the presence of specific information in the medication guide. | Databases | |
| Retrieve medication guide information based on the drug name. | Databases | |
| Retrieve the drug name based on the information provided in the patient package insert. | Databases | |
| Retrieve the patient package insert information based on the drug name. | Databases | |
| Retrieve drug names based on a specific ingredient present in the drug product. | Databases | |
| Retrieve a list of drug ingredients based on the drug name. | Databases | |
| Retrieve the SPL unclassified section information (Content not yet clearly categorized) based on the drug name. | Databases | |
| Retrieve the drug name based on the stop use information provided. | Databases | |
| Retrieve stop use information based on the drug name provided. | Databases | |
| Retrieve the drug name based on storage and handling information. | Databases | |
| Retrieve storage and handling information based on the drug name. | Databases | |
| Retrieve drug names based on the summary of safety and effectiveness information. | Databases | |
| Retrieve a summary of safety and effectiveness information based on the drug name. | Databases | |
| Retrieve drug names based on specific teratogenic effects categories. | Databases | |
| Retrieve teratogenic effects information based on the drug name. | Databases | |
| Retrieve drug names based on their use in specific populations, such as pregnant women, nursing mothers, pediatric patients, and geriatric patients. | Databases | |
| Retrieve information about the use of a drug in specific populations based on the drug name. | Databases | |
| Retrieve specific user safety warnings based on drug names. | Databases | |
| Retrieve drug names that have specific user safety warnings. | Databases | |
| Retrieve the drug names based on specific warning information. | Databases | |
| Retrieve warning information based on the drug name. | Databases | |
| Retrieve warnings and cautions information for a specific drug based on its name. | Databases | |
| Retrieve drug names based on specific warnings and cautions information. | Databases | |
| Retrieve information about side effects and substances or activities to avoid while using a specific drug. | Databases | |
| Retrieve the brand name and generic name from generic name or brand name of a drug. | Databases | |
| Retrieve information about all contraindications for use based on the drug name. | Databases | |
| Retrieve about the drug product’s indications for use based on the drug name. | Databases | |
| Get the drug’s generic name based on the drug’s generic or brand name. | Databases | |
| Find targets associated with a specific disease or phenotype based on efoId. | Databases | |
| Find diseases or phenotypes associated with a specific target using ensemblId. | Databases | |
| Explore evidence that supports a specific target-disease association. | Databases | |
| Retrieve warnings for a specific drug using ChEMBL ID. | Databases | |
| Retrieve the mechanisms of action associated with a specific drug using chemblId. | Databases | |
| Retrieve known drugs associated with a specific disease by disease efoId. | Databases | |
| Retrieve similar entities for a given disease efoId using a model trained with PubMed. | Databases | |
| Retrieve similar entities for a given target ensemblID using a model trained with PubMed. | Databases | |
| Find HPO phenotypes associated with the specified disease efoId. | Databases | |
| Find withdrawn and black-box warning statuses for a specific drug by chemblId. | Databases | |
| Get the count of entries in each entity category (disease, target, drug) based on a query string. | Databases | |
| Retrieve the efoId and additional details of a disease based on its name. | Databases | |
| Fetch the drug chemblId and description based on the drug name. | Databases | |
| Fetch indications (treatable phenotypes/diseases) for a given drug chemblId. | Databases | |
| Retrieve Gene Ontology annotations for a specific target by Ensembl ID. | Databases | |
| Fetch homologues for a specific target by Ensembl ID. | Databases | |
| Retrieve known target safety liabilities for a specific target Ensembl ID. | Databases | |
| Retrieve biological mouse models, including allelic compositions and genetic backgrounds, for a specific target. | Databases | |
| Retrieve genomic location data for a specific target, including chromosome, start, end, and strand. | Databases | |
| Retrieve information about subcellular locations for a specific target Ensembl ID. | Databases | |
| Retrieve synonyms for a specified target, including alternative names and symbols, using the given Ensembl ID. | Databases | |
| Retrieve tractability assessments, including modality and values, for a specific target Ensembl ID. | Databases | |
| Retrieve the target classes associated with a specific target Ensembl ID. | Databases | |
| Retrieve the Target Enabling Packages (TEP) associated with a specific target Ensembl ID. | Databases | |
| Retrieve interaction data for a specific target Ensembl ID, including interaction partners and evidence. | Databases | |
| Retrieve the disease ancestors and parents in the ontology using the disease EFO ID. | Databases | |
| Retrieve the disease descendants and children in the ontology using the disease EFO ID. | Databases | |
| Retrieve the disease’s direct location and indirect location disease terms and IDs using the disease EFO ID. | Databases | |
| Retrieve disease synonyms by its EFO ID. | Databases | |
| Retrieve disease description, name, database cross-references, obsolete terms, and whether it’s a therapeutic area, all using the specified EFO ID. | Databases | |
| Retrieve the therapeutic areas associated with a specific disease EFO ID. | Databases | |
| Retrieve significant adverse events reported for a specific drug ChEMBL ID. | Databases | |
| Get a list of known drugs and associated information using the specified ChEMBL ID. | Databases | |
| Get parent and child molecules of specified drug ChEMBL ID. | Databases | |
| Retrieve detailed information about multiple drugs using a list of ChEMBL IDs. | Databases | |
| Get drug name, year of first approval, type, cross references, and max clinical trial phase based on specified chemblId. | Databases | |
| Retrieve the synonyms associated with a specific drug ChEMBL ID. | Databases | |
| Retrieve the trade names associated with a specific drug ChEMBL ID. | Databases | |
| Retrieve the approval status of a specific drug ChEMBL ID. | Databases | |
| Retrieve chemical probes associated with a specific target using its Ensembl ID. | Databases | |
| Retrieve pharmacogenomics data for a specific drug, including evidence levels and genotype annotations. | Databases | |
| Get known drugs associated with a specific target Ensembl ID, including clinical trial phase and mechanism of action of the drugs. | Databases | |
| Retrieve the list of diseases associated with a specific drug ChEMBL ID based on clinical trial data or post-marketed drugs. | Databases | |
| Retrieve the list of targets linked to a specific drug ChEMBL ID based on its mechanism of action. | Databases | |
| Perform a multi-entity search based on a query string, filtering by entity names and pagination settings. | Databases | |
| Retrieve Gene Ontology terms based on a list of GO IDs. | Databases | |
| Retrieve genetic constraint information for a specific target Ensembl ID, including expected and observed values, and scores. | Databases | |
| Retrieve publications related to a target Ensembl ID, including PubMed IDs and publication dates. | Databases | |
| Retrieve publications related to a drug ChEMBL ID, including PubMed IDs and publication dates. | Databases | |
| Get the Ensembl ID and description based on the target name. | Databases | |
| Get disease EFO ID and description by disease name from OpenTargets. | Databases | |
| Get target Ensembl ID by target name. | Databases | |
| Get disease EFO ID by disease name. | Databases | |
| Find drug ChEMBL ID by drug name. | Databases | |
| Find targets associated with a specific disease or phenotype based on its name. | Databases | |
| Find diseases or phenotypes associated with a specific target. | Databases | |
| Explore evidence that supports a specific target-disease association. | Databases | |
| Retrieve warnings for a specific drug. | Databases | |
| Retrieve the mechanisms of action associated with a specific drug. | Databases | |
| Retrieve known drugs associated with a specific disease by disease name. | Databases | |
| Retrieve similar entities for a given disease using a model trained with PubMed. | Databases | |
| Retrieve similar entities for a given target using a model trained with PubMed. | Databases | |
| Find HPO phenotypes asosciated with the specified disease. | Databases | |
| Fetch indications (treatable phenotypes/diseases) for a given drug. | Databases | |
| Retrieve Gene Ontology annotations for a specific target. | Databases | |
| Fetch homologues for a specific target. | Databases | |
| Retrieve known target safety liabilities for a specific target. | Databases | |
| Retrieve biological mouse models, including allelic compositions and genetic backgrounds, for a specific target. | Databases | |
| Retrieve genomic location data for a specific target, including chromosome, start, end, and strand. | Databases | |
| Retrieve information about subcellular locations for a specific target. | Databases | |
| Retrieve synonyms for specified target, including alternative names and symbols. | Databases | |
| Retrieve tractability assessments, including modality and values. | Databases | |
| Retrieve the target classes associated with a specific target. | Databases | |
| Retrieve the Target Enabling Packages (TEP) associated with a specific target. | Databases | |
| Retrieve interaction data for a specific target, including interaction partners and evidence. | Databases | |
| Retrieve the ancestors and parents of a specific disease. | Databases | |
| Retrieve the descendants and children of a specific disease. | Databases | |
| Retrieve the locations of a specific disease. | Databases | |
| Retrieve synonyms for a specific disease. | Databases | |
| Retrieve the description of a specific disease. | Databases | |
| Retrieve the therapeutic areas associated with a specific disease. | Databases | |
| Retrieve chemical probes associated with a specific target. | Databases | |
| Get known drugs associated with a specific target, including clinical trial phase and mechanism of action of the drugs. | Databases | |
| Retrieve the list of diseases associated with a specific drug based on clinical trial data or post-marketed drugs. | Databases | |
| Retrieve the list of targets linked to a specific drug based on its mechanism of action. | Databases | |
| Retrieve genetic constraint information for a specific target, including expected and observed values, and scores. | Databases | |
| Retrieve diseases associated with a list of phenotypes or symptoms by a list of HPO IDs. | Databases | |
| Retrieve a phenotype or symptom by its HPO ID. | Databases | |
| Retrieve one or more HPO ID of a phenotype or symptom. | Databases | |
| Analyze protein sequence using InterProScan to identify functional domains, families, and GO terms. | Databases | |
| Search for similar protein sequences in UniProt Swiss-Prot database using BLAST. | Databases | |
| Retrieve the coefficient for the specified material. | Databases | |
| Get the magnetic moment value for an element. | Databases | |
| Lookup CAS number for a compound. | Databases | |
| Complete PubChem compound information. | Databases | |
| Search for similar compounds. | Databases | |
| Retrieve compound properties from PubChem. | Databases | |
| Resolve compound name to all identifiers. | Databases | |
| Analyze protein from UniProt. | Databases | |
| Get structure information. | Databases | |
| Get material price from compound. | Databases | |
| Fetches average price for multiple chemical substances identified by their CAS numbers. | Databases | |
| Query a SMILES and return their CAS numbers in string. | Databases | |
| Obtain lattice structure information from the provided MOF cif file name. | Databases | |
| Query a molecule name and return its SMILES string in Markdown format. | Databases | |
| Query a molecule name and return its CAS number in Markdown format. | Databases | |
| Search literature. | Literature Search | |
| Run the search engine with a given query, retrieving and filtering results. | Literature Search | |
| Run the search engine with a given query, retrieving and filtering results. | Literature Search | |
| Search PubMed for academic articles and retrieve abstracts. | Literature Search | |
| Retrieve publications related to a disease EFO ID, including PubMed IDs and publication dates. | Literature Search | |
| Retrieve publications related to a disease name, including PubMed IDs and publication dates. | Literature Search | |
| Retrieve publications related to a target, including PubMed IDs and publication dates. | Literature Search | |
| Retrieve publications related to a drug, including PubMed IDs and publication dates. | Literature Search | |
| Predict zero-shot sequence. | Model Services | |
| Predict zero-shot structure. | Model Services | |
| Predict protein properties. | Model Services | |
| Use the ESMFold model for protein 3D structure prediction. | Model Services | |
| Predict the ADMET properties of a molecule. | Model Services | |
| Given a protein sequence and its structure, employ the ProSST model to predict mutation effects and obtain the top-k mutated sequences based on their scores. | Model Services | |
| Retrieve similar entities for a given drug chemblId using a model trained with PubMed. | Model Services | |
| Retrieve similar entities for a given drug using a model trained with PubMed. | Model Services | |
| From compound name to ADMET prediction. | Model Services | |
| Predicts the solubility of a given protein sequence using a fine-tuned BioT5 model for the protein solubility prediction task. | Model Services | |
| Given a molecule SELFIES, generates its English description using a pre-trained T5 model. | Model Services | |
| Given a molecule description in English, generates its SELFIES and SMILES representation using a pre-trained T5 model. | Model Services | |
| Predicts whether a given molecule (SELFIES format) and a protein sequence can interact with each other and returns the result in Markdown format with a brief explanation. | Model Services | |
| Predicts whether two given protein sequences can interact with each other using a fine-tuned BioT5 model for the protein-protein interaction task with human dataset. | Model Services | |
| Predicts whether two given yeast protein sequences can interact with each other using a fine-tuned BioT5 model for the protein-protein interaction task with yeast dataset. | Model Services | |
| Predicts the solubility of a given protein sequence using a fine-tuned BioT5 model for the protein solubility prediction task. | Model Services | |
| Predicts the binary localization of a given protein sequence using a fine-tuned BioT5 model for the protein binary localization prediction task. | Model Services | |
| Manually adjust storage condition for a container. | Wet-lab Operations | |
| Apply acceleration (spin) to a container. | Wet-lab Operations | |
| Incubate a container at a location for a duration. | Wet-lab Operations | |
| Seal a container. | Wet-lab Operations | |
| Place a lid on a container. | Wet-lab Operations | |
| Perform washing of a target. | Wet-lab Operations | |
| Dry an item by a method. | Wet-lab Operations | |
| Close an item (e. | Wet-lab Operations | |
| Filter a sample through a filter type. | Wet-lab Operations | |
| Record data of a specified type. | Wet-lab Operations | |
| Let an item stand for some duration. | Wet-lab Operations | |
| Thaw materials at a specific temperature. | Wet-lab Operations | |
| Aspirate liquid from a source. | Wet-lab Operations | |
| Thermocycle a container with groups of steps. | Wet-lab Operations | |
| Wait for a specified duration. | Wet-lab Operations | |
| Heat a target to a temperature for a duration. | Wet-lab Operations | |
| Vortex an item. | Wet-lab Operations | |
| Separate phases/components from a target. | Wet-lab Operations | |
| Lyse a sample using a method and optional buffer. | Wet-lab Operations | |
| Add a component to a target container. | Wet-lab Operations | |
| Inoculate medium with inoculum. | Wet-lab Operations | |
| Inactivate a target using specified method and parameters. | Wet-lab Operations | |
| Anneal material at a temperature for a duration with optional cycles. | Wet-lab Operations | |
| Transfer material from source to destination. | Wet-lab Operations | |
| Resuspend a material/pellet in buffer. | Wet-lab Operations | |
| Mix components in a target container. | Wet-lab Operations | |
| Extract target material from a sample. | Wet-lab Operations | |
| Grind material with specified method and parameters. | Wet-lab Operations | |
| Prepare reagents or mixtures. | Wet-lab Operations | |
| Freeze an item at given temperature. | Wet-lab Operations | |
| Perform pipetting from source to destination. | Wet-lab Operations | |
| Perform extension (e. | Wet-lab Operations) | |
| Invert a container repeatedly to mix. | Wet-lab Operations | |
| Place an item at a location. | Wet-lab Operations | |
| Fill a container with a substance. | Wet-lab Operations | |
| Tap or flick a container to mix or settle. | Wet-lab Operations | |
| Centrifuge a container at a speed for a duration. | Wet-lab Operations | |
| Pellet material by specified method (e. | Wet-lab Operations) | |
| Aliquot a source into destination containers. | Wet-lab Operations | |
| Eliminate a target (e. | Wet-lab Operations) | |
| Measure a parameter on a sample. | Wet-lab Operations | |
| Balance items for equipment (e. | Wet-lab Operations) | |
| Shake a target container. | Wet-lab Operations | |
| Decant liquid from source to destination. | Wet-lab Operations | |
| Purify a sample using a method/kit. | Wet-lab Operations | |
| Perform sequencing run with specified parameters. | Wet-lab Operations | |
| Digest a sample with an enzyme under specified conditions. | Wet-lab Operations | |
| Elute material from a sample using a buffer. | Wet-lab Operations | |
| Stain a target with an agent. | Wet-lab Operations | |
| Discard items or materials safely. | Wet-lab Operations | |
| Dilute a sample by a factor. | Wet-lab Operations | |
| Dissolve material in a solvent under specified conditions. | Wet-lab Operations | |
| Abstract instruction to run an agarose gel and acquire an image using an appropriate DNA stain and imaging system. | Wet-lab Operations | |
| Generation experiment protocol given user prompt. | Wet-lab Operations | |
| Execute JSON with PCR operation server Params: json (str): executable JSON server_url (str): PCR operation server URL; default to Thoth-OP server URL Returns: exec_info (str): execution information Arg: arguments (dict) - Tool parameter dictionary | Wet-lab Operations |
The current list of all available SCP tools.
📊 논문 시각자료 (Figures)