Rare gastrointestinal lesions are infrequently encountered in routine endoscopy, restricting the data available for developing reliable artificial intelligence (AI) models and training novice clinicians. Here we present EndoRare, a one-shot, retraining-free generative framework that synthesizes diverse, high-fidelity lesion exemplars from a single reference image. By leveraging language-guided concept disentanglement, EndoRare separates pathognomonic lesion features from non-diagnostic attributes, encoding the former into a learnable prototype embedding while varying the latter to ensure diversity. We validated the framework across four rare pathologies (calcifying fibrous tumor, juvenile polyposis syndrome, familial adenomatous polyposis, and Peutz-Jeghers syndrome). Synthetic
Rare diseases affect over 300 million people worldwide, yet timely and accurate diagnosis remains challenging. This challenge is especially acute for rare gastrointestinal lesions in endoscopic practice, where many entities are documented by only a single image or a handful of cases [1][2][3]. Because endoscopic diagnosis is a dynamic, real-time process that relies on continuous multi-angle visual inspection and clinical experience rather than direct instrument readouts, limited exposure makes it difficult for clinicians to build reliable recognition of rare phenotypes. The same scarcity also leaves data-driven artificial intelligence (AI) models with too few examples to learn robust diagnostic patterns. Together, these factors increase the risk of missed or incorrect diagnoses [4][5][6][7].
Conventional augmentation is often insufficient for rare endoscopic lesions because it cannot reproduce the clinically meaningful appearance changes that shape diagnosis, including variations in lighting, viewing geometry, mucosal background, lesion scale, and fine surface texture. As a result, augmented images may increase quantity without providing the diversity that supports reliable recognition. Recent text-guided generative models, such as MINIM [8], can synthesize high-fidelity medical images across multiple organs and modalities and have been explored for improving downstream AI-assisted diagnosis via data augmentation. In endoscopic imaging, CCIS-Diff [9] generates controllable colonoscopy images using a diffusion-model prior. However, these approaches are difficult to apply to rare or sparsely documented lesions because they typically require substantial paired text and image data for effective adaptation, a requirement that is rarely met in extreme low-data settings.
To adapt generative models to a new lesion type when only a few examples exist, prior few-shot strategies commonly rely on concept-specific optimization, such as fine-tuning a pre-trained diffusion model or jointly optimizing prompts and model parameters [10][11][12]. In practice, maintaining a separate adapted model for each rare phenotype does not scale [13], and learning from a single reference image often overfits and reduces clinically acceptable variation. Training-free seed selection methods (e.g., SeedSelect [14]) can improve sample diversity, but their gains are often brittle across different prompts and clinical contexts. Prompt engineering approaches such as DATUM [15] may further increase diversity, yet they risk altering diagnostically critical morphology, which limits their suitability for rare disease synthesis.
Therefore, there is a pressing need for a cost-effective framework that can robustly adapt to and faithfully synthesize target rare lesions. We propose EndoRare, a retraining-free one-shot framework for endoscopic rare-lesion synthesis. EndoRare pretrains a medical-domain diffusion prior and integrates language-informed concept disentanglement to separate and control clinically meaningful attributes. During personalization, it freezes the diffusion backbone and learns (i) a compact prototype embedding from a single exemplar and (ii) encoder-derived, language-aligned visual concepts. These representations steer the denoising trajectory to produce diverse, clinically credible samples without any additional fine-tuning at inference.
Empirically, EndoRare achieves state-of-the-art performance on endoscopic datasets. In blinded clinician assessments of realism and class faithfulness, it consistently outperforms prior generative baselines. When used for data augmentation, EndoRare-generated samples lead to reliable improvements in AI-assisted diagnosis of rare lesions compared with conventional augmentation. To facilitate clinical adoption, exposure to EndoRare-generated cases also enhances trainees’ recognition of rare diseases, highlighting both translational and educational impact. 2 Results
Fig. 1 summarizes the EndoRare framework and evaluation design. EndoRare addresses the long-tail rarity in endoscopy that limits both clinical exposure and data-driven learning (Fig. 1a) through a three-stage pipeline (Fig. 1b): pretraining a knowledge-informed diffusion model on routine image-text pairs, learning an attribute-aligned image encoder, and deploying a controllable generator that combines a lesion-specific embedding with attributes extracted from real rare cases. Methodological details are provided in Section 4. We evaluate EndoRare along three axes (Fig. 1c) on four rare entities, CFT [16], JPS [17], FAP [18], and PJS [19] (Supplementary Table B1): (i) image fidelity and clinical faithfulness using quantitative metrics and blinded clinician ratings; (ii) downstream utility by training rare-lesion diagnostic models with EndoRare-augmented data against conventional augmentation and recent generative baselines; and (iii) clinical relevance via a within-subject pre-post reader study: novices were first trained using only one real anchor image per entity and then evaluated on a fixed mixed image set containing both rare and non-rare cases, where their task was to detect and identify rare lesions; they were subsequently retrained with additional EndoRare-generated exemplars and re-evaluated on the same mixed set.
We first evaluated the visual realism and clinical relevance of the synthesized images. As shown in Fig. 2a, EndoRare successfully reproduces critical endoscopic photometrics, such as mucosal sheen, specular highlights, and natural vignetting, while preserving the hallmark morphology of specific rare entities (e.g., solitary nodules for CFT/JPS versus carpet-like clusters for FAP/PJS). The variations in viewpoint, scale, and luminal background demonstrate that the model achieves substantial intra-class diversity without compromising class semantics.
To quantify this performance, we analyzed the trade-off between image fidelity and diversity (Fig. 2b). While DreamBooth achieved the lowest Fréchet Inception Distance (FID [20]: 253.43±28.02), it suffered from limited diversity (LPIPS [21]: 0.214±0.007). Conversely, the Zero Shot baseline maximized diversity (0.500 ± 0.002) but at the cost of significantly degraded fidelity (FID: 309.82 ± 29.49). EndoRare struck a favorable balance between these regimes, maintaining competitive fidelity (FID: 280.26 ± 11.64) while achieving high diversity (LPIPS: 0.489±0.021). In terms of semantic consistency (Fig. 2c), EndoRare (CLIP-I [22]: 0.847 ± 0.006) remained competitive with other generative baselines, ensuring that the synthesized diversity remains faithful to the text prompts.
Crucially, blinded assessments by expert gastroenterologists corroborated these objective metrics (Fig. 2d). In a study involving 700 randomly sampled images evaluated on a 3-point clinical scale, EndoRare achieved the highest overall rating (2.
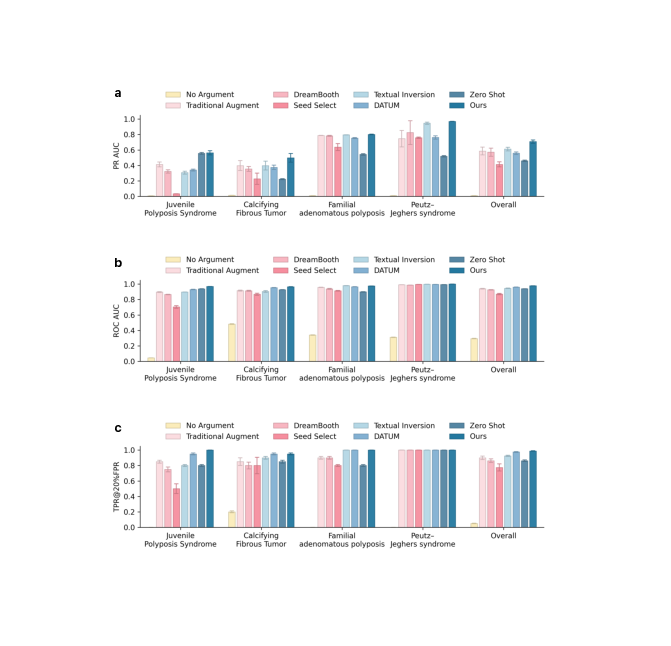
We evaluated the downstream utility of EndoRare by benchmarking a rare-lesion classifier trained with our augmented data against traditional augmentation and recent generative baselines. As summarized in Fig. 3a-c, incorporating EndoRare significantly enhances overall diagnostic performance. Specifically, our method raised the macroaverage PR-AUC to 0.708, surpassing the strongest baseline (Textual Inversion, 0.612) by a substantial margin (+0.096, p < 0.001). Similarly, EndoRare achieved the highest macro-average ROC-AUC of 0.978 and improved sensitivity at a clinically relevant operating point (TPR@20% FPR = 0.988), offering an absolute gain of 8.8% over traditional augmentation (p < 0.001). This performance advantage extends across individual disease categories. Endo-Rare yielded the most pronounced improvements in PR for challenging entities such as CFT (0.498 vs. 0.398 for Textual Inversion; p < 0.001). In terms of ROC-AUC, performance approached ceiling levels for several categories; notably, EndoRare led significantly on JPS (p < 0.001), CFT (p < 0.05), and PJS (p < 0.001). While performance on FAP was saturated across methods, EndoRare remained highly competitive (0.975), comparable to the top-performing baselines. Saturation effects were also observed for TPR@20% FPR, where EndoRare attained perfect scores (1.0) on JPS and FAP and tied for top performance on CFT (0.95).
Given that minimizing false positives is critical for reducing procedural burden in endoscopy, we further analyzed performance within the constrained low-FPR window (0-20%, Fig. 4). EndoRare yielded the highest macro partial AUC (pAUC) of 0.175, exceeding the strongest baseline (DATUM) by 5.8%. Entity-specific analysis confirmed that the largest gains occurred in CFT (∆ = +0.017), while PJS reached saturation (pAUC = 0.199). Although JPS performance was tied with Zero Shot in terms of pAUC, EndoRare demonstrated superior sensitivity at the 20% FPR operating point (0.95 vs. 0.80). Collectively, these results indicate that EndoRare-augmented training maintains higher cumulative sensitivity within the decision regions most relevant to clinical screening, preserving robustness across diverse rare entities.
To quantify the educational utility of EndoRare, we conducted a within-subject reader study assessing novice performance before and after exposure to synthetic cases (Fig. 5a). The results reveal a clinically important pattern (Fig. 5b). For entities with distinctive phenotypes (FAP and PJS), novice endoscopists were already near ceiling at baseline, indicating that these conditions are readily recognizable even with limited exposure. In contrast, for cryptic entities such as JPS and CFT, which are morphologically ambiguous and can be mistaken for routine benign polyps, synthetic training produced large gains. Before training, novices frequently missed these lesions, leading to near-zero sensitivity. After exposure to diverse synthetic exemplars, performance increased substantially (Table 1). For JPS, precision increased from 0.033 to 0.480 (∆ = 0.4467, p = 0.0052), with concurrent improvements in recall (∆ = 0.6500, p = 0.0004). Gains were even larger for CFT, where recall reached 1.000 (∆ = 0.9000) alongside an increase in precision (∆ = 0.8267, p = 0.0004). These results suggest that synthetic cases help trainees learn subtle visual cues that distinguish rare look-alike lesions from common findings. Importantly, improved sensitivity did not come at the cost of reduced precision. In the precision-recall plane (Fig. 6), trajectories for JPS and CFT move upward and rightward, consistent with simultaneous gains in recall and precision. This pattern indicates that synthetic training does not simply encourage broader labeling of rare conditions, but improves discrimination.
Performance on the distinctive categories (FAP and PJS) remained stable without degradation (p = 0.3739 for PJS), providing a safety signal that synthetic exposure does not induce confusion for conditions that are already visually recognizable. Together, these findings suggest that EndoRare can shorten the time needed to build diagnostic experience by providing concentrated exposure to rare, easily overlooked phenotypes. Full statistical details are provided in Supplementary Table B7.
To dissect the contribution of each architectural component, we performed an ablation in which PSE, TLE and FM were incrementally enabled and evaluated both qualitatively and quantitatively (Fig. 7a,b). Qualitatively (Fig. 7a), the full model most closely resembles the reference examinations: lesions retain rounded contours and stalks in pedunculated polyps as well as the lobulated, multi-nodule appearance in clustered cases, while global colour tone and illumination are better matched. The zero-shot baseline tends to preserve only coarse class cues and drifts at the instance level (shape and surface texture). Removing TLE introduces visible artefacts (e.g., radial banding near the endoscope rim) and over-smoothed textures.
As shown in Fig. 7b, starting from the configuration without any module (FID = 279.31; CLIP-I = 84.12; LPIPS = 0.4982), adding PSE reduces FID to 260.26 (-19.05; -6.8%) but also lowers CLIP-I to 81.48 (-2.64) and LPIPS to 0.3887 (-0.1095). See Supplementary Fig. A15
Clinical endoscopy faces chronic data scarcity for rare entities: novice readers and supervised classifiers alike see too few examples to learn stable decision rules. Our goal was therefore pragmatic and clinically motivated: to supply realistic and class-faithful exemplars that broaden exposure without imposing additional collection burdens or privacy risks. We framed this as one-shot synthesis for rare lesions, with two key desiderata: fidelity to lesion identity and diversity along clinically meaningful axes (e.g., color, location). We evaluated performance not only with image-similarity metrics but also with task-level utility and clinician judgment [23].
Conventional personalization by full fine-tuning (e.g., DreamBooth) can improve identity preservation but tends to shrink diversity and risks instance memorization under extreme data scarcity. EndoRare separates what to keep (lesion identity) from what to vary (non-critical attributes). Concretely, identity is recovered via prototypebased conditioning, and variability is injected by attribute-anchored text prompts, obviating any retraining of the diffusion model. This pipeline operates on prompts and prototypes rather than gradient updates over protected images, reducing both overfitting and privacy exposure relative to heavy-weight fine-tuning.
Our experiments show that EndoRare addresses the central challenge in image synthesis: balancing fidelity and diversity. As in Fig. 2b-c, it achieves substantially higher diversity (LPIPS 0.489 ± 0.021) while maintaining competitive fidelity (FID 280.26 ± 11.64) compared with established methods (Fine-tuned SD, DreamBooth), indicating avoidance of mode collapse without sacrificing anatomical realism. Its superior blinded clinical ratings (overall 2.32) for realism and class faithfulness further support clinical plausibility; the close performance in semantic alignment (CLIP-I 0.847 ± 0.006) indicates strong image-image correspondence without degrading visual quality.
Downstream results corroborate these findings (Fig. 3). Training with EndoRare augmentations improves PR-AUC (0.708 vs. 0.612 for the strongest baseline) and ROC-AUC (0.978 vs. 0.961), suggesting enhanced generalization rather than mere dataset inflation [24]. Gains are most pronounced where data are scarcest, as in CFT (+0.100), implying that EndoRare captures nuanced features of rare phenotypes that baselines miss. In reader studies, novices improved markedly on JPS (precision 0.033 → 0.480, F1 0.040 → 0.559) and CFT (precision 0.133 → 0.960, F1 0.114 → 0.978), indicating accelerated recognition of subtle pathological cues.
Because generic perceptual scores were developed for natural images, their relevance to endoscopy is limited: FID and related metrics can be insensitive to lesioncritical cues and rely on feature extractors misaligned with endoscopic content [25]. We therefore complemented them with blinded expert ratings as a clinical gold standard [26] and, beyond image-level assessment, with task utility via classification and reader studies aligned to real-world endpoints [27].
Recent work suggests that diffusion models, especially when fine-tuned with weak regularization, can memorize patient images and leak prototypes upon querying [28][29][30][31]. In our experiments, instance-level fine-tuned baselines (Fintune SD and DreamBooth) exhibited lower diversity at comparable fidelity [32], consistent with this risk profile. EndoRare mitigates memorization by avoiding gradient updates on patient pixels, enforcing a language-informed factorization, and sampling with attribute variation that discourages verbatim replication. Mitigation, however, is not proof of absence; future releases should include formal privacy audits (nearest-neighbour and attribute-leakage analyses, membership/attribute-inference tests), configurable safety margins (e.g., minimum attribute perturbations), and optional safeguards such as watermarking or differentially private training for auxiliary adapters [26,33,34].
This study has limitations. We evaluate four entities with five novice readers; larger multi-center studies across devices and expertise levels are needed to assess durability and generalizability [35]. Ceiling effects for some categories (FAP, PJS) limit detectable gains; future work should emphasize harder phenotypes and mixed-prevalence test sets to probe sensitivity-specificity trade-offs. Although clinical ratings and task-level endpoints improve interpretability beyond FID, the field lacks community-accepted, lesion-aware metrics (e.g., pathology-consistent feature tests, segmentation-or reportaligned consistency). The approach also assumes high-quality textual descriptors; robust report parsing and attribute validation from routine endoscopy reports are important next steps. Finally, while EndoRare reduces privacy exposure relative to fine-tuning, it does not eliminate it; standardizing privacy tests and red-team prompts in the release process should become required practice.
Taken together, carefully controlled, retraining-free synthetic augmentation can expand exposure to rare phenotypes for both humans and algorithms, improve classifier operating points at low false-positive rates, and offer a safer alternative to heavy fine-tuning that curbs instance memorization [36]. By prioritizing expert assessment and task utility alongside generic image scores, this work aligns generation with clinical value. In the near term, EndoRare can serve as a companion to reader education and model development in low-prevalence settings; in the longer term, the same disentanglement-and-control principles could support case-mix simulation, curriculum learning, and bias probing across institutions. Progress will hinge on standardizing clinical evaluations, auditing privacy systematically, and extending controllable generation to a broader spectrum of rare diseases and acquisition settings.
Task Definition. We define the task of one-shot rare disease image synthesis as follows. Let D S = {(I Si , R Si )} N S i=1 be source dataset of common disease images, where each image I Si ∈ R H×W ×3 denotes an RGB endoscopy image and R Si ∈ R is its associated text report. Notably, such source datasets are readily compilable from clinical records [37]. Given a single target image of rare disease D T = {I T }, our goal is to generate a synthetic set D G = {I j G } N G j=1 that preserves the unique characteristics of I T while ensuring sufficient fidelity and diversity. Diffusion Models. Diffusion Models (DMs) [38] generate images by progressively denoising a noisy sequence. Given an image I 0 , Gaussian noise is added incrementally, and a U-Net [39] ϵ θ (•, t) is trained to predict ϵ t at timestep t. Conditional generation incorporates text embeddings from a pretrained CLIP encoder τ θ (y) [22] via cross-attention (see Supplementary Fig. A14). We adopt Stable Diffusion [40], a Latent Diffusion Model (LDM) that operates in latent space for efficiency. The image is encoded as z = E(I 0 ) using a Variational Autoencoder [41], and the conditional LDM training objective is:
where z t is the noised latent at timestep t and y is a text condition.
As illustrated in Fig. 8, we first pretrain a text-to-image diffusion model on source datasets of common diseases. To steer learning toward clinically meaningful features, senior attending endoscopists re-annotate and enrich the original endoscopy reports, after which a large language model extracts salient attributes-morphology, color, pathology, and location-from these curated texts. We reorganize the extracted attributes into compositional prompts that provide a structured prior over typical lesion characteristics. Next, we introduce a cross-modal lesion-feature disentanglement module (Sec. 4.4) to align visual factors with their textual counterparts. After this concept-learning stage, given a single prototype image of a rare disease, we translate its disentangled visual factors into textual descriptions and combine them with learnable, prototype-specific embeddings to synthesize the final image.
To integrate domain knowledge from endoscopy reports in R, we use a Large Language Model (LLM) to extract four key attributes: morphology, color, pathology, and location. These attributes are formatted into a structured template: a polyp, color, and the pathology is , located at . The formatted text is encoded by a pretrained CLIP text encoder to obtain τ θ (y i ), which conditions the diffusion model. By optimizing the loss in Eq. ( 1) with these structured inputs, we embed domain-specific knowledge, enabling the model to capture meaningful lesion characteristics. This pretraining provides a broad representation of common disease appearances, ensuring generalization to rare lesions.
To disentangle intricate visual features of rare disease images, we introduce a crossmodal lesion feature disentanglement mechanism. This framework aligns attributespecific visual representations of I T with their corresponding textual embeddings.
For each endoscopy report R Si ∈ D S , we extract key textual attributes, denoted as Attr i . Each attribute attr k i ∈ Attr i is encoded using the CLIP text encoder τ θ , producing an embedding e k i = τ θ (attr k i ). Simultaneously, a dedicated branch processes the corresponding image I Si using a CLIP image encoder to obtain v k i . To enforce cross-modal consistency, we integrate an attribute alignment term into the LDM loss, optimizing the following objective:
where λ balances attribute alignment with the diffusion model’s reconstruction loss.
Here, y is the formatted textual description of R S as defined in Section 4.3.
Prototype-Specific Embedding. To generate rare disease images without retraining the diffusion model, we introduce a learnable vector p T , the Prototype-Specific Embedding, which captures the unique features of a target lesion I T . Given the noised latent sample z t at timestep t, the model ϵ θ is conditioned on p T to predict the noise. The Mean Squared Error (MSE) loss optimizes p T by comparing the predicted noise ϵ θ (z t , t, p T ) with the actual noise ϵ t :
L Recon = E z,p,ϵ∼N (0,1),t ϵ t -ϵ θ z t , t, p T 2 .
(
With the pretrained diffusion model ϵ θ fixed, adaptation occurs only in p T . To provide effective guidance, p T is initialized with the text embedding of the lesion class name from τ θ . Tailored Lesion Embedding. While p T reconstructs I T , the generated image I recon may lack diversity and realism. To address this, we leverage the cross-modal visual concept learning, where each CLIP-based encoder extracts attribute embeddings {v k T } from I T . These embeddings are formatted into the textual template (Section 4.3), forming the Tailored Lesion Embedding r T .
Embedding Fusion with Guided Denoising. To generate high-fidelity rare disease images, we fuse p T with r T , where p T retains core morphology and r T enhances diversity through attribute-based variations. The fusion process involves computing similarity matrices between r T and p T , deriving attention weights via softmax, and aggregating embeddings into the final fused representation e T , either by averaging or concatenation. The fused embedding e T then guides the denoising process: z t-1 = √ ᾱt-1 z t -√ 1ᾱt ϵ θ (z t , t, e T ) √ ᾱt + 1ᾱt-1 -σ 2 ϵ θ (z t , t, e T ) + σϵ, (4) where ϵ θ predicts noise given z t , e T , and timestep t. This iterative process continues until the fully denoised latent code z 0 is obtained and decoded by D(z 0 ) into the final endoscopic image.
Restrictions apply to the availability of the developmental and validation datasets. These datasets were used with participants’ permission for the present study and are not publicly available. De-identified data may be made available for research purposes from the corresponding authors upon reasonable request and subject to institutional and ethical approvals.
The code to reproduce the results is available at github.com/Jia7878/EndoRare.
This content is AI-processed based on open access ArXiv data.