Health system learning achieves generalist neuroimaging models
📝 Original Info
- Title: Health system learning achieves generalist neuroimaging models
- ArXiv ID: 2511.18640
- Date: 2025-11-23
- Authors: Akhil Kondepudi, Akshay Rao, Chenhui Zhao, Yiwei Lyu, Samir Harake, Soumyanil Banerjee, Rushikesh Joshi, Anna-Katharina Meissner, Renly Hou, Cheng Jiang, Asadur Chowdury, Ashok Srinivasan, Brian Athey, Vikas Gulani, Aditya Pandey, Honglak Lee, Todd Hollon
📝 Abstract
Frontier artificial intelligence (AI) models, such as OpenAI's GPT-5 (1) and Meta's DINOv3 (2), have advanced rapidly through training on internet-scale public data, yet such systems lack access to private clinical data. Neuroimaging, in particular, is underrepresented in the public domain due to identifiable facial features within MRI and CT scans, fundamentally restricting model performance in clinical medicine (3). Here, we show that frontier models underperform on neuroimaging tasks and that learning directly from uncurated data generated during routine clinical care at health systems, a paradigm we call health system learning, yields high-performance, generalist neuroimaging models. We introduce NeuroVFM, a visual foundation model trained on 5.24 million clinical MRI and CT volumes using a scalable volumetric joint-embedding predictive architecture. NeuroVFM learns comprehensive representations of brain anatomy and pathology, achieving state-of-the-art performance across multiple clinical tasks, including radiologic diagnosis and report generation. The model exhibits emergent neuroanatomic understanding and interpretable visual grounding of diagnostic findings. When paired with open-source language models through lightweight visual instruction tuning, NeuroVFM generates radiology reports that surpass frontier models in accuracy, clinical triage, and expert preference. Through clinically grounded visual understanding, NeuroVFM reduces hallucinated findings and critical errors, offering safer clinical decision support. These results establish health system learning as a paradigm for building generalist medical AI and provide a scalable framework for clinical foundation models.📄 Full Content
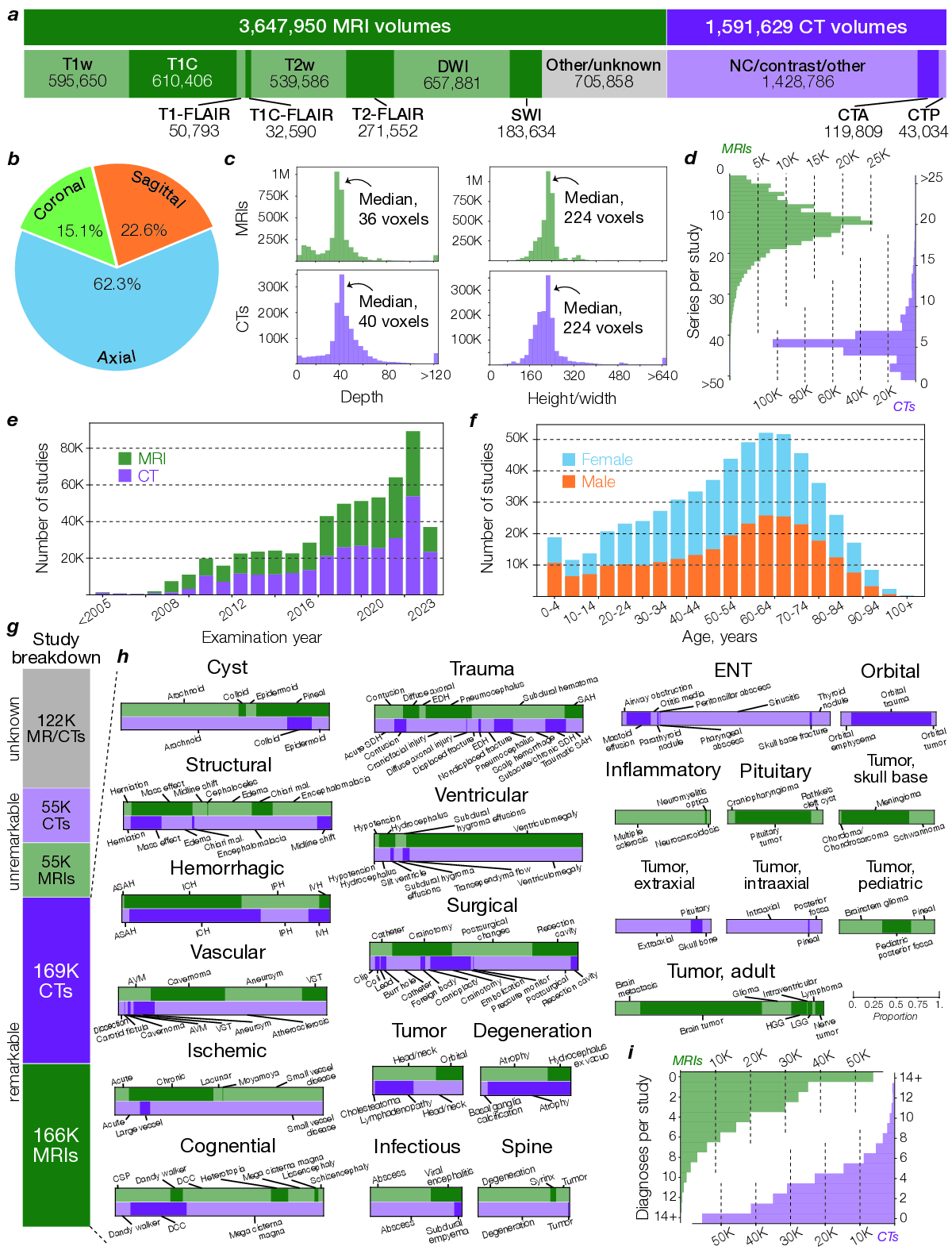
To compare pretraining on health system data with pretraining on public data, we conducted a year-long, multicenter diagnostic study across our health system. All patients consecutively evaluated between June 1, 2023, and May 30, 2024, who underwent a CT or MRI of the head or neck were enrolled without exclusion, yielding a representative sample of real-world clinical imaging. This cohort comprised more than 21,000 head CTs and 29,000 brain MRIs (Supplementary Data Table 1).
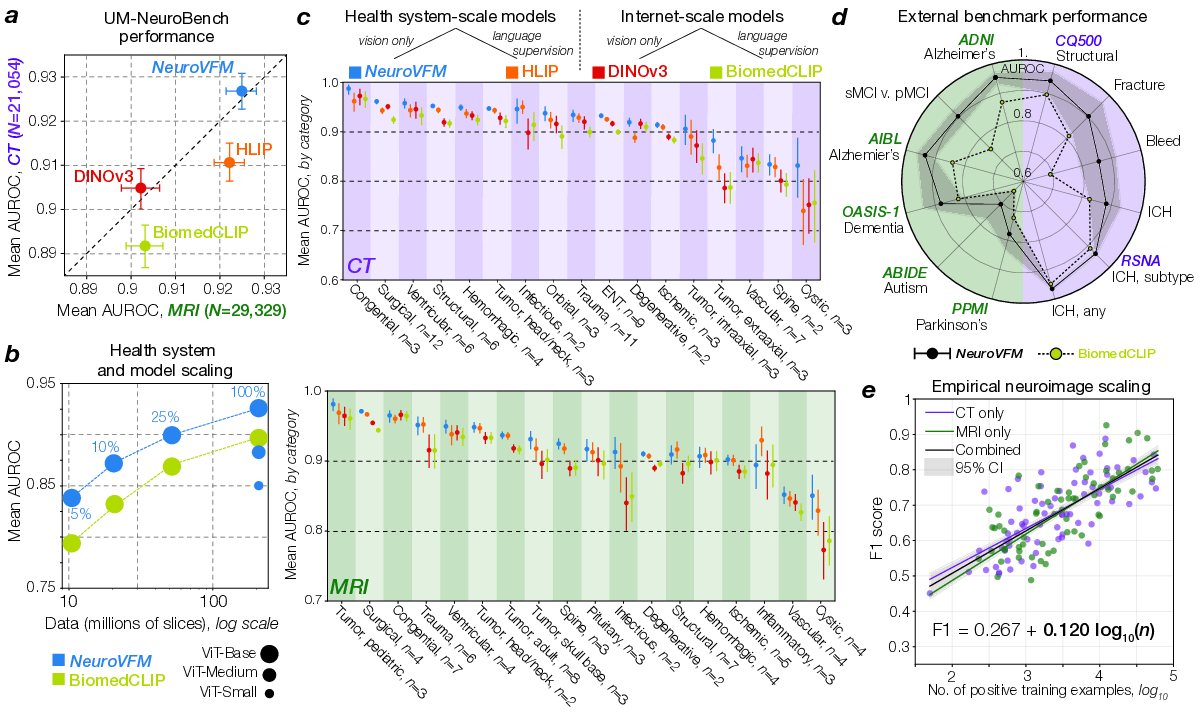
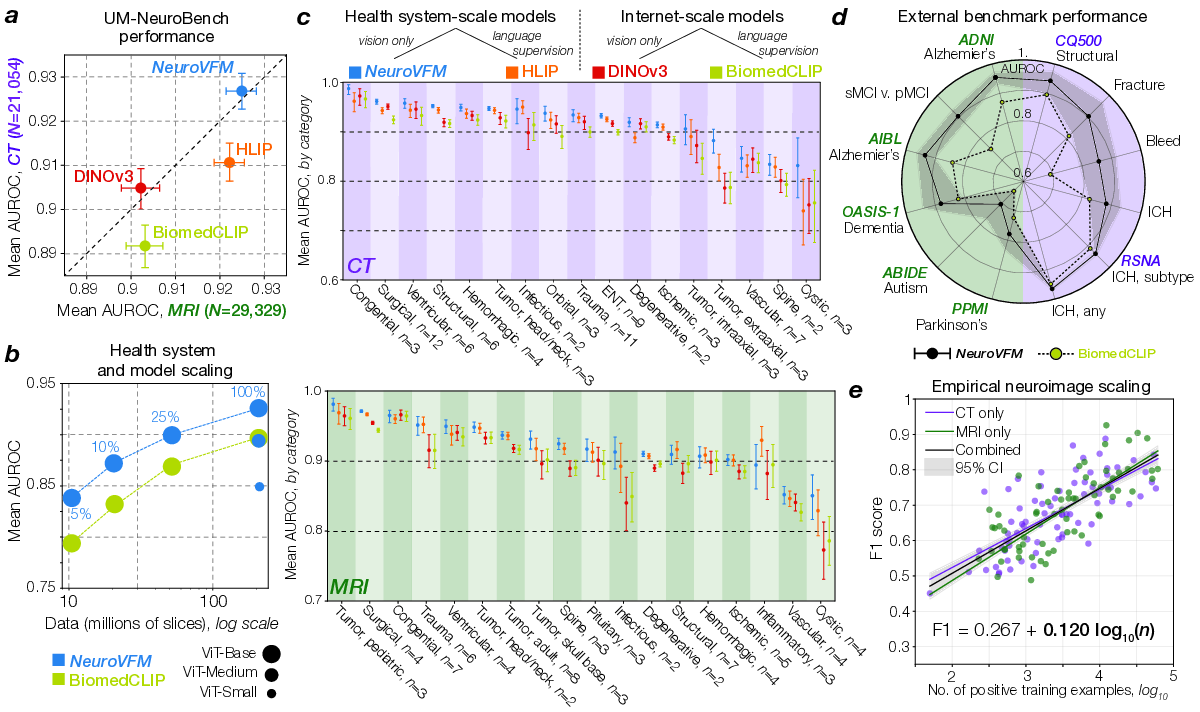
NeuroVFM achieved an average AUROC of 92.7 ± 0.4% across 82 CT diagnostic tasks and 92. 5 NeuroVFM outperformed internet-scale models by a wide margin. e, We discovered an empirical log-linear scaling relationship between the number of positive training examples and model performance. This relationship held across at least 4 orders of magnitude, imaging modalities, and models (Extended Data Fig. 4 and5). 0.4% across 74 MRI diagnostic tasks (Fig. 2a). We compared NeuroVFM with three state-of-theart models: the health system-trained HLIP model (13) and two internet-scale baselines, DINOv3 and BiomedCLIP. For each model, we trained an attentive pooling classifier on frozen embeddings from UM-NeuroImages. NeuroVFM outperformed all baselines for both CT and MRI diagnosis, demonstrating the value of scalable self-supervised learning and health system-scale pretraining. Its performance was comparable to or exceeded that of established diagnostic tools used by expert neuroradiologists such as the ASPECTS score for ischemic stroke (14) and the T2-FLAIR mismatch sign for diffuse glioma (15) (Extended Data Fig. 4 and5).
We found that NeuroVFM exhibits the hallmark properties of foundation models, with performance scaling predictably across both data volume and model size (Fig. 2b) (16). Empirically, we observed a non-saturating log-linear relationship between the number of positive diagnostic examples and performance metrics (Fig. 2e). NeuroVFM maintained stable performance across MRI manufacturers, magnetic field strengths, demographic subgroups, and medical centers (Extended Data Fig. 6). We further evaluated the model on multiple public neuroimaging benchmarks, including brain age estimation, dementia and autism classification, and intracranial hemorrhage detection. Across nearly all tasks, NeuroVFM outperformed internet-scale baselines by a wide margin (Fig. 2d, Extended Data Fig. 7).
Health system learning with Vol-JEPA gave rise to emergent abilities: clinically meaningful behaviors that were not explicitly supervised during training. Without segmentation or label supervision, NeuroVFM organized visual tokens into a latent neuroanatomic manifold that accurately encodes both spatial and semantic features of brain structures (Fig. 3a). Token-level encoding allows the model to match anatomic regions and pathologies across modalities, orientations, and imaging protocols, effectively performing zero-shot semantic matching (Fig. 3b). Similar to how token matching emerges in frontier models trained on natural images such as DINOv3, we observe analogous behavior in NeuroVFM. For example, the model recognizes that the pineal gland on coronal T2-weighted imaging corresponds to the same structure on sagittal T1-weighted post-contrast imaging. These results imply that NeuroVFM has learned an implicit brain atlas of human neuroanatomy. This emergent capability extends to zero-shot diagnostic transfer between MRI and CT, a property not observed in DINOv3 embeddings (Fig. 3c). , and all natural language processing metrics (d and Extended Data Fig. 9). e, Blinded clinical experts noted that NeuroVFM was more likely to generate correct key findings, less likely to hallucinate, and much less likely to make a laterality error compared to GPT-5. f, Clinical experts also preferred NeuroVFM reports more than 2:1 over GPT-5 reports. g-i, Illustrative examples of urgent, routine, and unremarkable head CTs and brain MRIs in the expert annotated testing set.
inherits robustness to variations in scanner hardware, protocols, and sites (Extended Data Fig. 6).
We define diagnostic grounding as the model’s ability to localize image regions that causally support a diagnostic prediction. To evaluate grounding, we used an attention-based multiple instance learning (AB-MIL) framework (see Methods) (17). NeuroVFM accurately localized pathologic regions and mapped them to corresponding radiologic diagnoses (Fig. 3d). For example, NeuroVFM correctly attends to and assigns the diagnostic labels to a patient with an epidural hematoma causing mass effect and midline shift. This comprehensive visual understanding extends across diverse pathologies, as illustrated in Extended Data Fig. 8. NeuroVFM also produced explainable classifier predictions for complex diseases such as Alzheimer’s, attending to mesial temporal structures, including the hippocampus and amygdala, known to be implicated in disease progression (Fig. 3e) (18). These results parallel the emergent abilities seen in frontier foundation models (19), suggesting that healthsystem-scale self-supervised training can yield qualitatively new capabilities in medical AI.
Frontier models such as GPT-5 and Claude Sonnet 4.5 are generative vision-language systems trained at internet scale on a vast corpus of public medical images and text. We tested whether NeuroVFM, when paired with an open-source language model trained only with autoregressive next-word prediction, could outperform these frontier models on the task of generating radiology reports for clinical triage.
Triage is a high-yield application for AI, benefiting patients, radiologists, and clinicians through increased efficiency and early prioritization of urgent findings (20,21). In this task, the model must generate itemized reports for real-world, uncurated, clinical, neuroimaging studies that accurately describe all key pathologic findings to classify each study into one of three clinical categories: unremarkable, routine, or urgent (Fig. 4a).
The frozen NeuroVFM model was paired with the open-source language model Qwen3 in a standard LLaVA-1.5-style visual instruction tuning framework to evaluate the quality of NeuroVFM’s learned representations for report generation (Fig. 4) (22,23). Our goal was not to optimize for report generation performance but to assess the feasibility of training a simple generative model directly from NeuroVFM embeddings. The vision-language model was trained using standard supervised fine-tuning on the UM-NeuroImages dataset to generate all key pathologic findings (Extended Data Fig. 9). We used frontier reasoning models GPT-5 and Claude Sonnet 4.5 as baselines, prompting each to generate key findings (Supplementary Data Fig. 5). A fixed prompt and language model (GPT-5 or Sonnet 4.5) were then used as standardized judges to assign triage levels based on the generated reports, providing a standardized and model-agnostic evaluation protocol (Supplementary Data Fig. 6). All models were tested on 300 expert-selected CT and MRI studies, balanced across modality and triage level.
NeuroVFM outperformed GPT-5 and Claude Sonnet 4.5 on both three-tier triage accuracy and detection of urgent findings (Fig. 4b,c). When compared with ground-truth radiology reports, NeuroVFMgenerated findings achieved higher scores across all natural language evaluation metrics, including METEOR and ROUGE (Fig. 4d and Extended Data Fig. 9). In blinded expert evaluation, the key finding error rate of NeuroVFM-generated reports was roughly half that of GPT-5 (approximately 10% versus 20%), with substantially fewer hallucinations and laterality errors. An international cohort of three blinded clinical experts preferred NeuroVFM-generated reports more than two to one over GPT-5-generated reports with high inter-rater concordance (Fig. 4f; Fleiss’ 𝜅 = 0.718).
Figure 4g-i shows representative NeuroVFM-generated reports across urgent, routine, and unremarkable triage levels. NeuroVFM accurately identified large mass lesions, including intraparenchymal hematomas and infiltrative tumors, and recognized associated findings such as midline shift, effacement of the basal cisterns, and brain herniation to determine lesion severity. The model also distinguished intracranial from extracranial hematomas, such as scalp hematomas, an essential distinction for triaging traumatic brain injury. NeuroVFM correctly identified non-urgent findings such as arachnoid cysts, even when producing mild mass effect, and consistently triaged unremarkable CT and MRI studies across imaging protocols and age groups, including those with high-risk clinical indications such as “worst headache of life” or “acute stroke.” We observed that both NeuroVFM and GPT-5 appropriately used the clinical indication to improve triage performance, whereas Claude Sonnet 4.5 struggled when given images alone (Extended Data Fig. 9). Examples spanning the full range of pathologies encountered in routine clinical practice are shown in Extended Data Fig. 10.
Health systems are knowledge bases and data engines that capture the collective experience of public health and clinical medicine. Here, we demonstrate that directly experiencing the clinical world through health system learning achieves high-performance neuroimaging foundation models. These task-agnostic vision models, pretrained on large and diverse clinical datasets, provide robust and transferable representations for downstream tasks. By integrating domain-specific intelligence learned from private clinical data with general-purpose reasoning, health system learning and NeuroVFM provides a roadmap towards AI systems that interpret and act on clinical information with subspecialist depth, expert reliability, and safe deployment.
Frontier models benefit from the vastness of publicly available information; however, this breadth limits their depth in technical domains, such as medicine (24). Internet data rarely reflects the complexity and diversity of real-world clinical imaging, where disease manifestations, acquisition protocols, and patient anatomy can vary widely. Health system learning represents a fundamentally different paradigm (25,26). Rather than learning from descriptions of the clinical world or curated hand-selected patient examples, health system learners, like NeuroVFM, experience the world itself, modeling the raw signatures of diseases embedded in unfiltered clinical data. World models have gained attention as a strategy to address the limitations of sequence modeling and reinforcement learning (27)(28)(29). NeuroVFM shows that intelligence can emerge from observing clinical practice itself.
Our long-term vision for NeuroVFM is to complement frontier models, not replace them in medicine. Agentic AI systems, including GPT-5 and Claude Sonnet 4.5, have integrated tool use to improve performance in specific domains, such as mathematics, science, and medicine (30-32). We foresee NeuroVFM and other health system models being integrated into agentic AI systems as external modules that provide a grounded understanding of clinical data. Frontier models will be able to reason over structured, clinically calibrated outputs from NeuroVFM, reducing failure modes and providing an auditable substrate for action. We believe expert-level performance across most cognitively complex domains will require the integration of domain-specific modules into general-purpose AI models.
Limitations and directions for future research include the need to integrate temporal and additional multimodal data streams, including pathology, genomics, and longitudinal clinical outcomes, to construct unified representations of disease progression. While the present study focuses on neuroimaging, the unified Vol-JEPA architecture is inherently extensible to other medical imaging modalities and body parts. Finally, while NeuroVFM exhibits emergent interpretability, translating these insights into actionable clinical interfaces will require new methods for human-AI collaboration, uncertainty quantification, and prospective evaluation in clinical workflows.
In summary, NeuroVFM demonstrates that generalist medical intelligence can be built from the health system itself. Learning directly from health system data yields representations that translate to diagnosis, triage, and report generation. We provide a scalable blueprint towards the development of medical foundation models that will be transformative in 21 st century healthcare.
We view health system learning as the environment within which generalist medical foundation models can emerge (33). To clarify the concept and differentiate it from existing foundation model training methods, such as internet-scale or life science training, we elaborate on several principles that define health system learning.
-
Directly learning the clinical world: Models should learn from data generated during routine patient care with no or minimal data processing or curation, experiencing the breadth and complexity of clinical medicine. This ensures robustness, realism, and coverage of clinically important edge cases.
-
Scalable training: Health system datasets require scalable supervision. Self-supervised learning objectives, including predictive, contrastive, and autoencoding objectives, are the current preferred strategy. However, other methods, such as semi-supervision or energy-based objectives, are candidates for scalable health system learning.
-
Clinical grounded representations: Medical foundation models trained through health system learning should internalize anatomy, pathology, and physiologic variation that supports accurate diagnosis, cross-modal reasoning, and interpretable grounding, thereby reflecting the epistemic structure of clinical expertise.
-
Multimodal medical learning: Health system data are multimodal representations of a single underlying object: the patient. Any single modality is a partial and incomplete view of the patient.
Multimodal models can achieve more complete representations and better disentangle explanatory features across modalities 5) Integration with agentic and multimodal AI systems: Health system learning is inherently modular. The resulting domain-specific foundation models can serve as expert visual or multimodal modules within larger agentic systems. This complements general-purpose models, enabling clinically safe, domain-aligned reasoning at scale.
Radiology studies at large academic medical centers are stored in picture archiving and communication systems (PACS). We queried the University of Michigan PACS via a SQL-based interface (Sectra Data Warehouse) for neuroimaging studies that satisfy: 1) an acquisition date prior to June 1, 2023; 2) an examination of body part(s) including the head, brain, orbits, face, or neck; and 3) a modality of MRI or CT. The specific SQL query parameters are in Supplementary Data Figure 1.
The initial query yielded 645,989 unique studies.
To ingest the full breadth of health system data without manual curation, we applied an automated pipeline to standardize intensities and dimensions. Series were resampled to 1 × 1 × 4 mm (4 mm along the acquisition axis from DICOM) and saved as 8-bit. MRI intensities were clipped to the 0.5-99.5th percentile before quantization. CT volumes yielded three windows: brain (width=80, level=40; 8-bit), subdural/blood (width=200, level=80; 4-bit), and bone (width=2800, level=600; 4-bit). 4-bit windows reduced storage while preserving contrast for key pathologies. Background masks were derived via Otsu and Hounsfield thresholding for MRI and CT, respectively. During training, volumes were cast to float, scaled to [0,1], and mean-normalized using statistics derived from the training set. Extended Data Fig. 1b summarizes this pipeline.
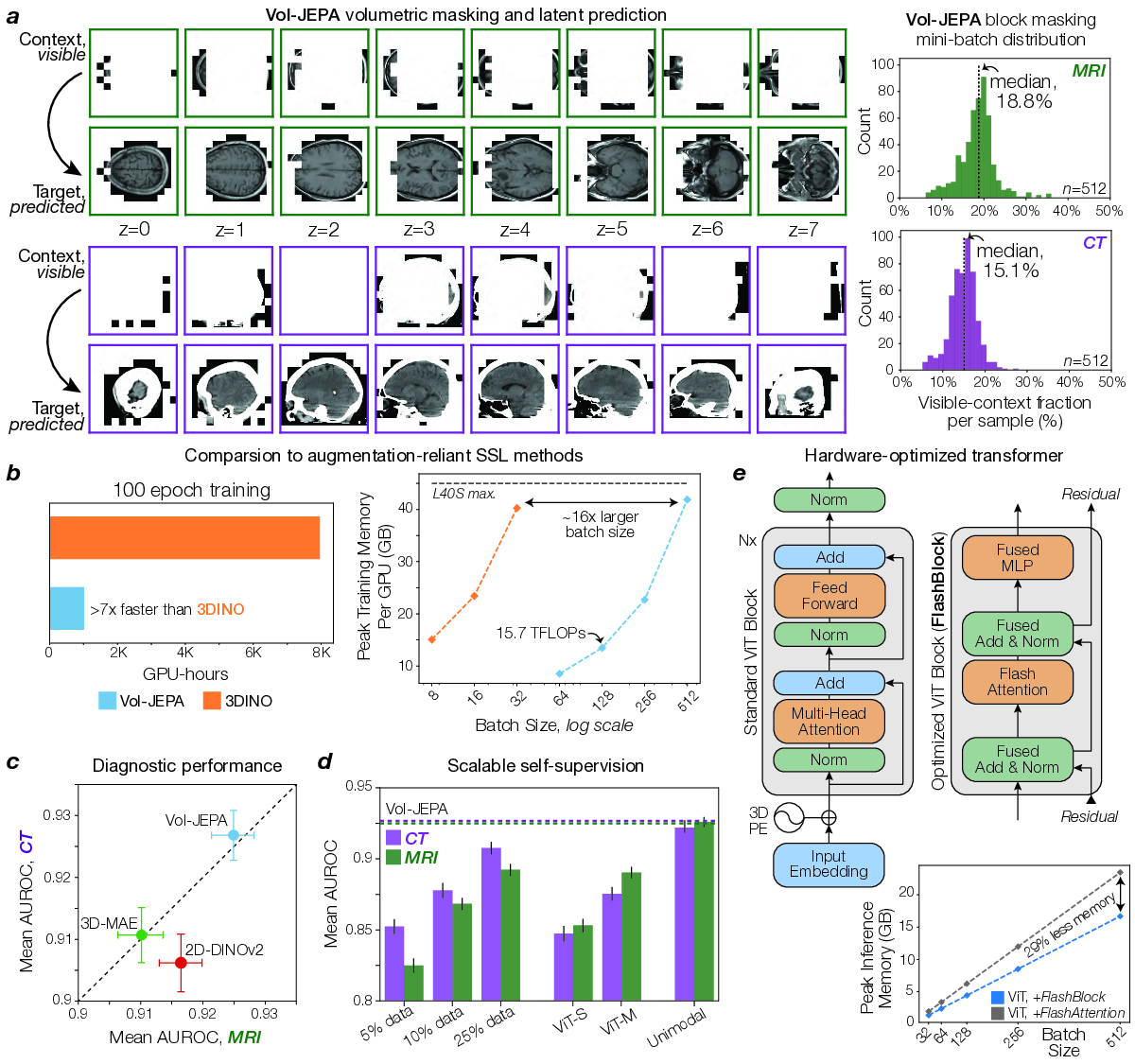
The core training objective for NeuroVFM was large-scale self-supervision, applied at the individual volume level, across the entire UM-NeuroImages dataset. NeuroVFM employs a joint-embedding predictive architecture (JEPA) to optimize a masked modeling objective directly in the representation space. First, input volumes are tokenized into non-overlapping 3D patches of 4×16×16 voxels. Each 3D volume is then partitioned into a small, visible context region, 𝑥, and larger, masked target region, 𝑦. The tokenized context region is processed by a trainable student encoder, 𝐸 𝜃 . The resulting context representations 𝐸 𝜃 (𝑥), along with positional information of the masked target patches, are then fed to a predictor module, 𝑃 𝜑 , which generates predictions for the representations of these target patches.
The complete set of tokens from the input volume (representing both context and target regions) is processed by a teacher encoder, whose weights 𝐸 θ are updated as an exponential moving average (EMA) of the student encoder’s weights after each step and are not directly optimized by gradient descent. The training objective minimizes the difference between the predicted representations of the target patches and those generated by the teacher network for the same target patches. This is formulated as:
where Δ 𝑦 is a set of learnable tokens that represents the masked target patches and sg(•) denotes a stopgradient operation. This representation-level objective is computationally efficient and encourages the learning of semantic features without requiring explicit voxel-level augmentations or decoding.
Volumetric-JEPA (Vol-JEPA) extends the principles of I-JEPA (8) and V-JEPA (10) for self-supervision to volumetric neuroimages, predicting representations of masked 3D target patches based on visible 3D context patches. This encourages the model to learn the shared anatomy of the brain, head, and neck. The masking strategy was foreground-focused: context and target patches were sampled exclusively from the pre-computed head mask. Masking was performed in two ways: (1) multiple large crops are sampled, with their union forming the masked target, and (2) a small crop is sampled as context, with the complement being the target. A random subset of context patches is then dropped, serving as additional target patches to predict. Unlike approaches that truncate sequences within a mini-batch to a uniform length, our implementation leverages FlashAttention-2 (37) to directly encode variablelength sequences of context and target patches derived from input volumes. To ensure robustness to varying patient orientations, we applied random axis permutations and flips to each volume, using the same transform for both student and teacher inputs. A hyperparameter search indicated optimal context region sampling ratios (from the total patches within a given crop) of 25% for MR volumes and 20% for CT volumes, with a patch dropout rate of 20%. An overview of this masking strategy can be found in Extended Data Fig. 3a.
During training, volumes were truncated along each axis to a maximum of 20 patches to bound the token count per crop. At inference, the same encoder was applied to the entire volume without truncation, leveraging its ability to handle variable-length token sequences. For CT scans, which are typically viewed with multiple windowing presets (e.g., brain, subdural, bone), we implemented a weighted sampling strategy during training. For pretraining, one of these three CT window settings was randomly selected and applied with probabilities of 0.7 (brain), 0.15 (subdural), and 0.15 (bone), and all windows were used during inference. Full training details are in Supplementary Data Table 6.
Most visual grounding evaluation requires an object detection module to output a bounding box around an image region given a label or text prompt. This is not feasible for the current study because there are no object detection datasets or models sufficiently powerful to evaluate neuroimage grounding on the scale and complexity of UM-NeuroImages. Because most neurologic pathologies are spatially small relative to the full study, study-level labels provide only weak supervision. We leveraged an attention-based multiple instance learning (AB-MIL) framework to assess neuroimage grounding (17). AB-MIL is known to assign high attention to diagnostic regions in medical images (38). Unfortunately, the standard AB-MIL framework, with its ‘aggregate-then-classify’ design, cannot resolve a critical grounding ambiguity: it is unable to disentangle a patch’s importance, where the model looks, from its directional contribution, why it is considered positive or negative evidence.
We address this gap with a pooling operation that reverses the standard order to a ‘classify-thenaggregate’ model. For a task with 𝐾 classes and a bag of 𝑁 instances, we first compute classification logits for each instance 𝑖 using a multilayer perceptron (MLP), 𝜓 𝑝 . A separate attention MLP, 𝜓 𝑚 , generates 𝐾 class-specific attention scores per instance. For each class, these scores are normalized with a softmax across instances to obtain class-specific attention weights. The final bag-level logits, 𝑝(𝑥), are the sum of the element-wise product of the per-instance logits and their corresponding attention weights:
Here, 𝑓 (𝑥 𝑖 ) is the frozen feature vector for instance 𝑖, 𝛼 𝑖 is the vector of class-specific attention weights, and • denotes the Hadamard product. This formulation yields interpretable, label-specific attention maps that reflect both the importance and directional contribution of each region to the diagnostic decision, providing a scalable means of evaluating grounding without region-level annotations.
We evaluated NeuroVFM against two families of baselines: (1) Internet-scale pretrained encoders, and
(2) methods directly trained on UM-NeuroImages. All backbones were frozen and evaluated with the same study-level attentive probe and data splits. Full architecture details, training hyperparameters, and evaluation configurations can be found in Supplementary Data Table 6 and our GitHub repository.
A schematic of the study-level pooling strategies can be found in Extended Data Fig. 1e.
We chose two baselines representative of the dominant Internet-scale paradigms: For 2D encoders, 3D volumes were processed slice-wise according to the model’s respective preprocessing pipeline (e.g., DINOv3 necessitates resizing to 224 × 224 and normalizing with ImageNet mean/std), with slice-level features aggregated to produce study-level predictions.
To provide a controlled comparison of representation learning methods, we trained key architectures on our UM-NeuroImages dataset. For comparison to vision-only self-supervision strategies, we benchmarked against a 3D Masked Autoencoder (VideoMAE) (40) with an 85% foreground-aware masking ratio, and DINOv2 model ( 41) trained on 2D neuroimaging slices derived from UM-NeuroImages volumes. The DINOv2 features were aggregated in a similar fashion to the pretrained 2D vision encoders. For vision-language alignment comparison, we trained HLIP (13), a state-of-the-art 3D medical vision-language model, on our 444,188 study-report pairs and evaluated its study image encoder. HLIP training minibatches were balanced across MRI and CT studies, and the CLIP objective was computed within each modality to prevent trivial cross-modality discrimination.
The UM-NeuroImages dataset contains two decades of clinical neuroimaging data from a large academic health system, spanning a broad spectrum of neurological presentations. To evaluate imaging-only diagnostic ability, we evaluated NeuroVFM on a prospective, temporally held-out test cohort beginning immediately after the retrospective window. Labels were derived from radiology reports using a LLM extraction pipeline, with a subset manually verified by clinical experts. We defined a clinically organized ontology of 74 MRI and 82 CT diagnoses (Extended Data Fig. 2h). Encoders were frozen, and modality-specific attentive probes were trained for multi-label prediction using class-weighted binary cross-entropy. Hyperparameters and class-specific thresholds were selected on the validation set. We report per-class AUROC, balanced accuracy, sensitivity, and specificity on the test cohort, with 95% CIs from study-level bootstrap resampling (10,000 replicates). Full results are provided in Supplementary Data Table 2 and3.
We studied performance along data scale, model capacity, and modality with resource-normalized protocols. For modality, we held the optimization budget fixed at ∼510K training steps and compared (i) a single multimodal model vs. (ii) two unimodal models (one MRI, one CT) trained separately. For data and model scaling, we varied (a) the fraction of UM-NeuroImages (5%, 10%, 25%), and (b) the encoder size (ViT-Small, ViT-Medium, ViT-Base). Larger backbones (e.g., ViT-Large) were left to future work due to computationally prohibitive hyperparameter search.
We quantified how per-task performance scales with the amount of supervision. For each diagnostic class 𝑐 and modality 𝑚 ∈ {CT, MRI}, we calibrated model probabilities on the validation set using Platt scaling and computed F1 on the held-out test set. We excluded classes with fewer than 30 training positives (𝑛 pos ) and 10 testing positives. We fit a simple linear model of F1 vs. log 10 (𝑛 pos ) via OLS with HC3 SEs. Modality fixed effects and interactions assessed CT vs. MRI differences. Encoder comparisons (NeuroVFM vs. HLIP, DINOv3, and BiomedCLIP) used ANCOVA to test slope equality. If interaction terms were non-significant, a shared slope was used, otherwise encoder-specific slopes were retained. Label-equivalence was reported as the fold-increase in positives a baseline requires to match NeuroVFM at a fixed F1, with 95% CIs from 10,000 bootstrap replicates over classes.
To assess the out-of-distribution generalization, we evaluated frozen NeuroVFM performance on eight public neuroimaging benchmarks. For all tasks, we trained an attentive probe without updating the encoder. Within each dataset, we held out 20% of subjects as a stratified test set. On the remaining 80%, we performed 8-fold iterative stratified cross-validation to select probe hyperparameters. The 8 probes trained on the cross-validation folds were then used to generate logits on the held-out test set, which were averaged per study to form the final ensemble. All reported metrics include 95% subject-level percentile CIs (10,000 bootstrap replicates). This approach rigorously tests the quality of the learned representations and, through the probe’s attention weights, allows for the identification of class-discriminative tokens for each task.
We evaluated NeuroVFM on six public MRI datasets, spanning a range of neurological and psychiatric conditions. On the multi-site OpenBHB dataset (42), we benchmarked brain age regression to test for fine-grained anatomical representation. For dementia-related tasks, we utilized the ADNI dataset (43) to perform CN vs. Alzheimer’s Disease (AD) classification and to distinguish progressive from stable Mild Cognitive Impairment (sMCI vs. pMCI), with a 20% stratified test set held out within ADNI. We then evaluated the ADNI-trained AD classifier externally on the OASIS-1 (44) and AIBL (45) datasets, applying the frozen probe without further fine-tuning to distinguish cognitively normal individuals (CDR = 0) from those with dementia (CDR ⩾ 1). We further evaluated diagnostic classification on several consortium datasets: differentiating individuals with Autism Spectrum Disorder from typically developing controls on the ABIDE dataset ( 46) and Parkinson’s Disease from healthy controls on the PPMI dataset (47).
Performance on detecting critical neuroradiological findings was evaluated on two public non-contrast head CT cohorts, selected to test generalization on both a large-scale challenge dataset and a smaller, deeply-annotated clinical cohort. The large-scale 2019 RSNA-ICH Challenge dataset (48) was used to benchmark multi-label classification of intracranial hemorrhage and its five subtypes (epidural, intraparenchymal, intraventricular, subarachnoid, and subdural). The high-quality, expert-annotated CQ500 dataset (49) was used for a more extensive evaluation across 14 diagnostic labels, including detailed hemorrhage characterization (subtypes, laterality, chronicity), skull fracture detection, and signs of structural abnormality (e.g., mass effect and midline shift).
To test whether NeuroVFM’s frozen representations align with neuroanatomy, we paired unsupervised feature analyses with reference neuroanatomical segmentations, which were derived from SynthSeg (12). First, we examined the low-dimensional organization of patch embeddings by running t-SNE on features from representative neuroimages and annotating each point by the majority SynthSeg region (frontal, parietal, temporal, occipital, insula, hippocampus, brainstem, and cerebellum) within its patch. We additionally tested whether NeuroVFM learns an approximate shared anatomical coordinate across subjects, modalities, sequences, and orientations via a patch-matching task. For each query patch from a neuroimage 𝐴, we searched over all patches in a different neuroimage 𝐵 (different subject and acquisition) by L2-normalizing features and identifying the nearest neighbor by cosine similarity. An imaging expert defined a set of canonical neuroanatomical regions (e.g., sella, pineal region, torcula), and we counted a match when both patches lay within the same expert-defined region in their respective scans, without any explicit spatial alignment or supervision. This effectively tests for zero-shot anatomical registration capabilities emergent from the self-supervised objective. Finally, we evaluated whether coarse neuroanatomy emerges without supervision by clustering dense, overlapping patch embeddings computed via sliding-window inference. A k-means model (𝑘 = 3) fit on embeddings from the initial window defined canonical clusters that were propagated to all windows and combined into a high-resolution map by voxel-wise majority vote. To identify the brain parenchyma cluster, we computed the IoU between each cluster mask and the SynthSeg brain mask, ultimately selecting the cluster with the highest IoU. Performance was summarized as the mean highest IoU across 30 neuroimages (10 each of axial, sagittal, and coronal natively) from the PPMI dataset, chosen as an external dataset with the full head and neck tissues included.
To evaluate NeuroVFM’s potential as a visual backbone for multimodal applications, we adapted a LLaVA-1.5-style visual instruction-tuning framework (23). This experiment was designed not to optimize report generation performance, but rather to assess the feasibility of coupling NeuroVFM’s learned representations with a large language model (LLM) with minimal architectural modifications.
NeuroVFM-LLaVA comprises three components: (1) the frozen NeuroVFM visual encoder, ( 2) an open-source LLM (Qwen3-14B) (22), and (3) a connector module to bridge them.
Standard LLaVA connectors use a 2-layer MLP to project visual features into the LLM’s word embedding space. This approach is insufficient for the high dimensionality of multi-sequence neuroimaging, where a single study may comprise a large and variable number of visual tokens (⩾20K). To address this, our connector module first employs a perceiver that operates sequence-wise (e.g., on T1, T2, FLAIR independently) (50,51). The perceiver aggregates the variable-length token sequence from each scan into a fixed-length representation of 64 latents. These fixed-length latents (total latents = 64 × 𝑛𝑢𝑚_𝑠𝑒𝑞𝑢𝑒𝑛𝑐𝑒𝑠) are then concatenated and passed to a 2-layer MLP projector.
The training dataset was derived from 444,188 unique neuroimaging studies. To create the text pairs, original radiology reports were summarized using GPT-4.1-mini to extract a concise list of key radiological findings.
Acknowledging that naive training on imbalanced medical datasets can degrade performance, we curated the training set via data resampling. Using the previously extracted diagnostic labels from each report, we performed weighted random sampling with replacement, assigning each study a weight inversely proportional to the prevalence of its rarest associated label. This process, which aims to balance the representation of less common pathologies, resulted in the final training dataset of ∼270K unique image-text pairs (444,188 total including duplicates).
Following the LLaVA methodology, training proceeded in two stages.
- Stage 1 (Connector Pre-training): Only the connector module (perceiver and MLP) weights are updated. The model was trained to map NeuroVFM image features to the corresponding reference key findings, which were formatted as a single, concatenated string. 2. Stage 2 (Full Fine-tuning): Both the connector and the LLM weights are updated. In this stage, the model was trained on an instruction-following task. The input prompt was fixed to: “Generate a concise report of the key positive findings for this study.” The target output was the same set of findings, but formatted as a structured JSON list ordered by clinical relevance.
Further details on training hyperparameters are provided in Supplementary Data Table 7.
We evaluated NeuroVFM-LLaVA against two proprietary multimodal frontier models: GPT-5 (gpt-5-2025-08-07, “reasoning” medium, “verbosity” low) and Claude Sonnet 4.5 (claude-sonnet-4-5-20250929, “thinking” enabled). This evaluation assessed two criteria: (1) the factual accuracy of generated findings and (2) the clinical utility of these findings for a downstream triage task. To support this evaluation, we established Business Associate Agreements (BAAs) with vendors offering commercially available HIPAA-compliant access to frontier models, specifically OpenAI and Anthropic. This enabled the secure exchange of protected health information (PHI) while preserving patient privacy.
A primary challenge in benchmarking against proprietary models is that they accept only 2D images and have limitations on total input image tokens. To create a fair comparison with our 3D-native model, we developed a systematic 3D-to-2D conversion pipeline.
- All 3D volumes (preprocessed with the NeuroVFM pipeline) were converted into 224 × 224 pixel 2D slices by resampling along the axis of acquisition and keeping every other slice. 2. Non-diagnostic and derived sequences (e.g., scout images, phase/magnitude maps) were excluded. 3. Due to API token limits, inputs were constrained to 15 sequences per study for GPT-5 (∼360 slices) and 4 for Claude Sonnet 4.5 (∼96 slices). 4. If a study exceeded this limit, sequences were deterministically prioritized based on clinical relevance (i.e., for MRI: post-contrast T1, DWI/ADC, FLAIR, SWI, T2, non-contrast T1; for CT: at least one brain, blood, and bone window). 5. All selected slices were converted to float and normalized between [0,1], and slices with >90% black pixels were dropped.
All generative models were evaluated on “UM-NeuroImages-Triage”, a new, manually-curated test set of 600 studies (300 validation, 300 holdout) derived from our prospective test set. This set was hand-selected by neuroimaging experts to be balanced across three acuity classes (“Unremarkable”, “Routine”, “Urgent”) and two modalities (MRI, CT).
We assessed report quality through automated metrics, BLEU-2 (52), ROUGE-L (53), and METEOR (54), as well as with human evaluation. To ensure effective blinding, all model outputs were standardized to a uniform text format. For NeuroVFM-LLaVA and GPT-5, a neuroimaging expert (T.H.) performed a structured review of each study to quantify (1) capture of key findings, (2) clinically significant hallucinations, and (3) laterality errors. To evaluate preferences, three neuroimaging experts from the United States and Europe (R.J., A.K.M., T.H.) then performed a blinded, randomized pairwise review comparing generated findings from NeuroVFM-LLaVA and GPT-5 against the ground truth clinical report. For each case, model outputs were randomly labeled “Report A” and “Report B”. Evaluators selected their preferred report (“Report A”, “Report B”, or “Both”) based on overall clinical utility for triaging. Because Claude Sonnet 4.5 exhibited substantially lower triage accuracy on UM-NeuroImages-Triage, we restricted expert review and preference testing to NeuroVFM-LLaVA and the strongest baseline (GPT-5).
To quantify the clinical utility of the generated findings, we employed an “LLM-as-a-judge” pipeline. In each analysis, we designated a frontier model (GPT-5 or Claude Sonnet 4.5) as a separate “triager LLM”, instructed with a strict, pre-defined set of criteria (Supplementary Data Fig. 6). We first confirmed that each triager achieved high accuracy when classifying acuity from the ground-truth radiology report alone. The triager LLM was then prompted to classify each study as “Unremarkable”, “Routine”, or “Urgent” based solely on the generated text findings from each model (NeuroVFM-LLaVA, GPT-5, and Claude Sonnet 4.5) and the clinical indication. Using GPT-5 vs. Claude Sonnet 4.5 as the triager yielded comparable results, indicating that the protocol is model-agnostic and not biased toward any single frontier model. This procedure converts report generation into a 3-class classification problem and provides a quantitative measure of clinical utility.
Large language models were used to assist with code prototyping and manuscript editing. All analyses, text and code were reviewed and verified by the authors, who take full responsibility for the work.
Our research was approved by the University of Michigan Institutional Review Board (HUM00229133).
All MRI and CT data were acquired under secondary data usage. The methods were carried out in accordance with the IRB’s guidelines, regulations, and policies. All human subjects that met inclusion criteria as stated above were included in the study.
( Extended Data Figure 1 | Extended NeuroVFM workflow. a, We queried the Michigan Medicine PACS for neuroimaging studies of the head, brain, face, orbits, and neck. After removing non-image and corrupted entries, the UM-NeuroImages cohort comprises 566,915 CT and MRI studies, of which 444,188 have paired radiology reports. Per-modality counts of volumes, studies, and reports are shown. To obtain training and evaluation labels at scale, we used an LLM report-parsing pipeline ( 7) that ( 1) extracts a list of expert-defined diagnoses and (2) converts free-text impressions to structured, itemized findings. Examples of both outputs are shown. b, Raw CT/MRI volumes are first resampled to 1 × 1 × 4 mm (4 mm along the native acquisition axis) to reflect conventional clinical slice spacing. MRI intensities are clipped to 0.5-99.5th percentile, and CT volumes are windowed into brain, blood, and bone using standard Hounsfield windows. All volumes are quantized to 8-bit (CT blood/bone further reduced to 4-bit) to reduce footprint. Modality-specific foreground masks remove air only (no skull-stripping). The resulting dataset contains 11.8 billion 3D 4x16x16 patches (approximately 9 TB). c, Example CT/MRI volumes after background removal and tokenization. For Vol-JEPA training, volumes are partitioned into context-target pairs. Mask placements and lengths are randomized to cover diverse spatial configurations. d, In Vol-JEPA, a student volumetric transformer first encodes the context, after which a predictor combines these latents with the masked target positions to predict target latents. A teacher encoder (an EMA of the student) processes the full volume to produce target latents for the masked region. The objective minimizes the smooth L1 distance between the predicted and teacher target latents. Only the student and predictor receive gradients, and ultimately the student is used for downstream inference. e, To compare fairly across architectures, we use a unified study-level attentive probing strategy across all available tokens. 2D models (e.g., DINOv3 and BiomedCLIP) encode each slice, with classification, register, and patch tokens from all slices fed to the study-level probe. 3D volume-level models (e.g., NeuroVFM and VideoMAE) encode each volume, with all tokens pooled jointly. 3D study-level models (e.g., HLIP) natively output study classification tokens, but for parity we use all available output tokens in the study-level probe. f, We used NeuroVFM to condition a multimodal LLM in a LLaVa-1.5 style to generate preliminary neuroimaging findings. First, each volume in a study is encoded with frozen NeuroVFM. The resulting tokens are condensed by a per-volume Perceiver resampler to 64 latents. These then pass through the LLM connector (two-layer MLP) and are fed as vision tokens to a decoder-only LLM (e.g., Qwen3-14B). During finetuning, the clinical indication is given as text input and the model is trained to generate the itemized findings from a. An example indication and generated findings are shown. g, Illustrative deployment of NeuroVFM within a secure hospital environment. Each time a patient undergoes neuroimaging, an internal function call applies NeuroVFM to generate preliminary findings. An external state-of-the-art reasoning model receives only these findings (no images or PHI) to perform tasks such as triage and referral. Its output can then trigger service-line alerts (e.g., “page neurosurgery”). Extended Data Figure 4 | Extended CT diagnostic results. CT diagnostic evaluation comprises 82 diagnoses defined on UM-NeuroImages. All encoders were frozen and evaluated using the same study-level attentive probe (Extended Data Fig. 1e). a, Logit-label performance matrix for the NeuroVFM CT classifier, where each cell is AUROC of logit 𝑖 against ground-truth label 𝑗 (main diagonal shows per-diagnosis AUROC). Rows and columns are reorganized by hierarchical consensus clustering (bootstrapped similarity, average linkage). Lower left, reference co-occurrence matrix computed from ground truth using min-normalization,
. b, Following the empirical scaling observed in Fig. 2e, we fit a shared slope across NeuroVFM and three baselines (BiomedCLIP, DINOv3, and HLIP) via ANCOVA (𝑝 ≪ 0.05), relating calibrated F1 score to log 10 of the number of positive training instances. Intercept shifts yield data-equivalence factors (how many more positives a baseline requires to match NeuroVFM at a fixed F1), shown with 95% CIs. c, Cross-modal transfer results for seven diagnoses with shared semantics across MRI/CT. An MRI-trained classifier is evaluated zero-shot on CT, and we plot ΔAUROC = (MRI-on-CT) -(CT-on-CT) with 95% bootstrap CIs. For NeuroVFM, all differentials lie within the a priori equivalence band of ±0.05 (green), whereas DINOv3 exhibits consistent modality shift. d, We compared NeuroVFM results on two representative CT detection tasks (top, acute ischemic stroke; bottom, aneurysmal subarachnoid hemorrhage) to published studies. NeuroVFM achieves AUROC comparable to expert neuroradiologists (14,55) and to validated AI systems (56,57). For prior studies, AUROC was approximated as the mean of the reported sensitivity and specificity. Extended Data Figure 5 | Extended MRI diagnostic results. MRI diagnostic evaluation comprises 74 diagnoses defined on UM-NeuroImages. All encoders were frozen and evaluated using the same study-level attentive probe (Extended Data Fig. 1e). a, Logit-label performance matrix for the NeuroVFM MRI classifier, where each cell is AUROC of logit 𝑖 against ground-truth label 𝑗 (main diagonal shows per-diagnosis AUROC). Rows and columns are reorganized by hierarchical consensus clustering (bootstrapped similarity, average linkage). Lower left, reference co-occurrence matrix computed from ground truth using min-normalization,
. b, Following the empirical scaling observed in Fig. 2e, we fit a shared slope across NeuroVFM and three baselines (BiomedCLIP, DINOv3, and HLIP) via ANCOVA (𝑝 ≪ 0.05), relating calibrated F1 score to log 10 of the number of positive training instances. Intercept shifts yield data-equivalence factors (how many more positives a baseline requires to match NeuroVFM at a fixed F1), shown with 95% CIs. c, Cross-modal transfer results for seven diagnoses with shared semantics across MRI/CT. A CT-trained classifier is evaluated zero-shot on MRI, and we plot ΔAUROC = (CT-on-MRI) -(MRI-on-MRI) with 95% bootstrap CIs. For NeuroVFM, all differentials lie within the a priori equivalence band of ±0.05 (green), whereas DINOv3 exhibits consistent modality shift. d, We compared NeuroVFM results on two representative MRI differentiation tasks (top, brain metastasis vs. high-grade glioma; bottom, low-vs. high-grade glioma) to published studies. For NeuroVFM, the score was the pairwise logit margin (logit 1 -logit 2 ), and AUROC was computed on this margin. NeuroVFM achieves AUROC comparable to expert neuroradiologists (15,58) and to validated AI systems (59,60). For prior studies that only report sensitivity and specificity, we approximate the AUROC as the average. Extended Data Figure 6 | Subgroup and health system analysis. Descriptive statistics for the prospective testing corpus and robustness of NeuroVFM across common sources of health system variation. a, Composition by scanner manufacturer and MRI field strength. b, Composition by age/sex and ethnicity. c, Composition by acquisition site within the health system. We limited the analysis to the three most common sites: University of Michigan (UM) Main, West, and East. d, Per-diagnosis robustness checks by (left) manufacturer and (right) MRI field strength. For each eligible cell (subgroup × diagnosis; requires at least 10 positives and negatives present), we compute the AUC differential relative to the full test set (Δ 𝑔 = AUC 𝑔 -AUC all ) using 10,000 paired, study-level bootstrap resamples to form percentile 95% CIs. Points show Δ 𝑔 , and the vertical dashed lines mark an a priori equivalence band of ± 0.05 AUROC. The y-axis summarizes CI separation index, 𝑆 = Δ 𝑔 half-width 95% , where 𝑆 > 1 indicates the 95% CI excludes 0 (i.e., a significant deviation). Cells with 95% CIs entirely within ± 0.05 AUROC were interpreted as showing no material deviation. Most diagnoses remain within the equivalence band, with outliers annotated (CT, purple; MRI, green). e, Mean AUROC (± 95% CIs) for MRI vs. CT stratified by age/sex, ethnicity, and site. No material differences present for age/sex and ethnicity. At the site level, MRI performance is lower at the East site, while CT is comparable across all. Extended Data Figure 7 | Open benchmarks performance. All encoders (NeuroVFM, blue; HLIP, orange; DINOv3, red; BiomedCLIP, olive) were frozen and evaluated using the same study-level attentive probe (Extended Data Fig. 1e). Error bars show 95% CIs. a, Brain age estimation results on the OpenBHB dataset (T1 MRIs). Scatter of RMSE (x-axis) vs. MAE (y-axis) per model shown. NeuroVFM is competitive (MAE, 2.805 years), second to BiomedCLIP. Strong 2D baselines likely benefit from dense slice-level features available to the probe. b, Intracranial hemorrhage detection and localization results on the 2019 RSNA-ICH dataset (non-contrast CT). Left, F1 for ICH detection and each subtype (IPH, IVH, SDH, SAH) shown. Right, PR lift for EDH (AUPRC/prevalence; EDH, 1.88%). NeuroVFM matches or exceeds other baselines across all tasks. c, We used three Alzheimer’s/dementia cohorts to test whether features generalize beyond a single study: ADNI (training, with a held-out test set) and two independent test sets (AIBL and OASIS-1). ADNI to AIBL reflects classic domain shift, whereas ADNI to OASIS-1 probes robustness as OASIS-1 provides a dementia severity score (CDR 0 vs. ⩾1). Left, training on ADNI and testing on AIBL/OASIS-1 yields strong AUPRC for NeuroVFM (DINOv3 comparable). Right, the same ADNI-trained NeuroVFM classifier exhibits high AUROC on the held-out ADNI test set and transfers well to AIBL and OASIS-1. These results indicate the learned NeuroVFM features capture disease-relevant information rather than site-specific cues, consistent with Fig. 3e where attention concentrates on structures known to be discriminative for Alzheimer’s disease (18). d, Correct laterality assignment is clinically important yet rarely tested. The CQ500 dataset provides expert “Bleed-Left/Bleed-Right” labels, allowing targeted laterality check given correct bleed detection. For each model, among studies it correctly flags as bleed, we horizontally flip the volume and measure changes in both the Bleed-Left and Bleed-Right logits (i.e., Δ 𝐿 = logit flipped 𝐿 logit 𝐿 , Δ 𝑅 = logit flipped 𝑅 logit 𝑅 ). We summarize performance as AUROC over Δ 𝐿 and Δ 𝑅 against the ground-truth side. NeuroVFM is near perfect, representative of strong anatomical encoding. DINOv3, on the other hand, underperforms, plausibly due to strong flip-invariance from augmentations. e, K-means clustering (k=3) of dense patch embeddings from NeuroVFM yields coarse tissue-wise clusters that recover brain parenchyma without supervision. See Methods for full description. f, We quantitatively validated attention maps of models’ UM-NeuroImages classifier against ground-truth segmentations on the BraTS21 (‘brain tumor’), ATLAS v2.0 (‘acute ischemic stroke’), and OpenMS (‘multiple sclerosis’) datasets. Specifically, we computed whether the voxel with the highest attention lies in the ground-truth segmentation, known as pointing game accuracy. Result and a representative example for each dataset are shown, with NeuroVFM demonstrating the highest degree of visual grounding. g, Schematic outlining how the predictor of NeuroVFM can be visualized via kNN reconstruction. To probe what the predictor infers for a masked target region, we project predicted latents back to image space via 𝑘-nearest-neighbor (kNN) retrieval. First, we build a patch latent databank by encoding patches from a reference set with the EMA teacher, yielding latent keys 𝑧 𝑖 and storing their corresponding image patches 𝑥 𝑖 . For a held-out volume, we encode a context region with the student encoder and use the predictor to produce latent predictions ẑ 𝑗 for each masked target patch. For each ẑ 𝑗 , we perform kNN search (cosine distance) over {𝑧 𝑖 }, retrieve the top-𝑘 patches, and form a pseudo-reconstruction x 𝑗 = 1 𝑘 𝑖∈ 𝑁 𝑘 𝑥 𝑖 . h, Example rollout of a FLAIR volume from BraTS21. Here, three axial slices were passed to the student as context and the rest of the sequence was predicted and rolled out. Qualitatively, the reconstructions recover tumor extent and surrounding anatomy from the correct neuroimaging sequence and rollouts across adjacent slices maintain structural consistency, indicating that the predicted latents are accurate. This example illustrates the challenge of interpreting real-world clinical neuroimaging studies that do not fit neatly into singular diagnoses, which is often observed in standardized research datasets.
NeuroVFM was able to recognize the postsurgical and post-traumatic changes, while both GPT-5 and Claude Sonnet 4.5 were not able to interpret this image correctly. c, Classic subarachnoid hemorrhage pattern due to a ruptured anterior communicating artery aneurysm. d, Contrasted CTs are much less common. NeuroVFM was able to both recognize that this was a contrasted CT and interpret that the findings were most consistent with metastatic brain disease. e, A common example of restricted diffusion due to an acute infarct located in the right posterior cerebral artery (PCA) distribution. Supplementary Data Figure 1 | SQL query to retrieve neuroimaging studies. Using the SDW, we retrieved all CT and MRI studies whose body part or description contained “brain,” “head,” “orbit(s),” or “neck,” returning exam metadata and report text. The SQL query filters studies acquired before June 1 2023 for model development. The held-out prospective set applies the same query on and after that date.
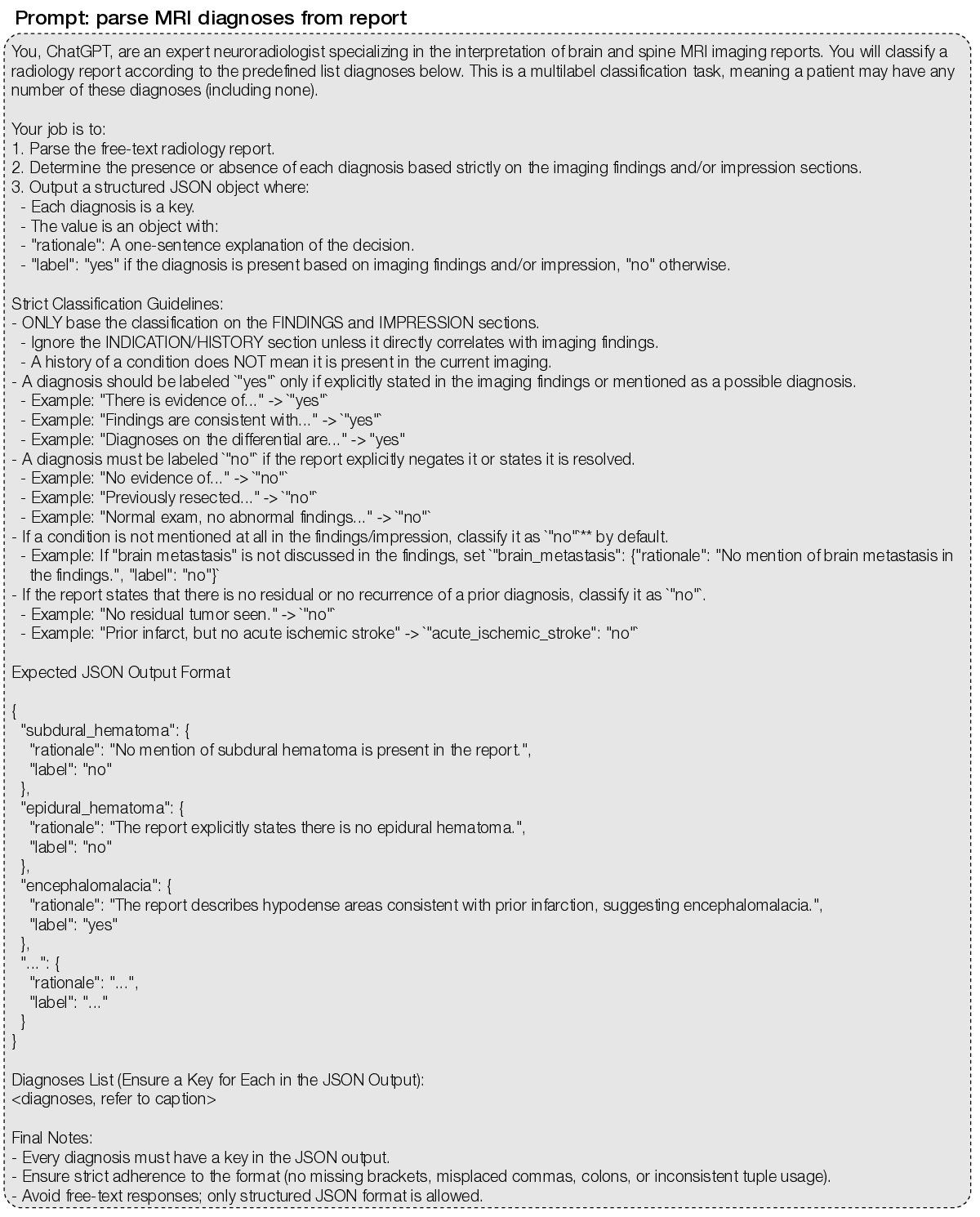
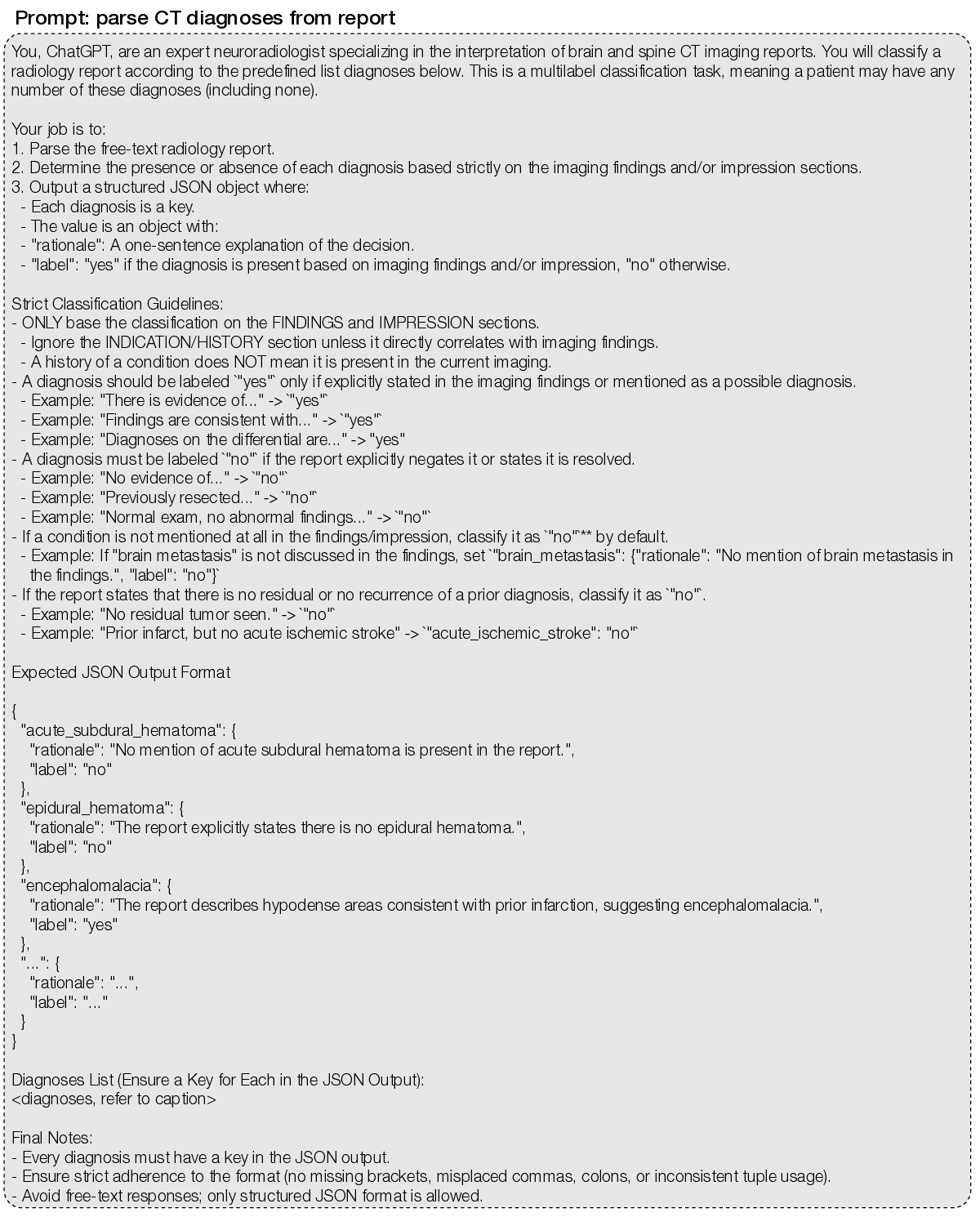
Supplementary Data Figure 2 | MRI diagnosis extraction prompt. We used an LLM-based annotation pipeline (7) with GPT-4.1-mini to convert free-text radiology reports into structured labels for 74 expert-defined MRI diagnoses. The prompt requires, for each diagnosis, a present/absent label and a brief rationale supporting the decision. The full list of MRI diagnoses is: subdural_hematoma; epidural_hematoma; brain_contusion; diffuse_axonal_injury; cerebral_aneurysm; arteriovenous_malformation; cavernous_malformation_cavernoma (“yes” if radiologist confident for cavernoma; “no” if possible microbleed); venous_sinus_thrombosis; acute_ischemic_stroke (“yes” for acute ischemia/infarct with diffusion restriction); chronic_ischemic_stroke (“yes” for chronic or evolving ischemia/infarct without diffusion restriction); moya_moya_disease; small_vessel_ischemic_disease (also known as microvascular ischemic disease); lacunar_stroke; intracranial_hemorrhage (“yes” for any intracranial hemorrhage, excluding microhemorrhage); intraparenchymal_hemorrhage (“yes” for hemorrhages within the brain, usually spontaneous and due to hypertension); intraventricular_hemorrhage; aneurysmal_subarachnoid_hemorrhage (“yes” if due to ruptured aneurysm or other vascular cause); traumatic_subarachnoid_hemorrhage (“yes” if due to head trauma); multiple_sclerosis; neuromyelitis_optica; neurosarcoidosis; brain_abscess (“yes” if radiologist is confident for brain abscesses or subdural empyemas); viral_encephalitis (most commonly herpes encephalitis involving the temporal and frontal lobes); arachnoid_cyst; pineal_cyst; epidermoid_cyst (“yes” for intracranial epidermoid cyst only); colloid_cyst; brain_tumor (“yes” for any intracranial mass, growth, or neoplasm); orbital_tumor (“yes” for any orbital mass, growth, or neoplasm); head_neck_tumor (“yes” for tumor/cancer in the oral cavity, throat, larynx, nasal cavity, salivary, and thyroid gland); spine_tumor (“yes” for any spine mass, growth, or neoplasm); nerve_tumor (“yes” for nerve, nerve root, or nerve sheath tumors); spinal_degenerative_changes; glioma (“yes” if mostly likely primary glial tumor); high_grade_glioma (“yes” if MRI features for high-grade glioma); low_grade_glioma (“yes” if MRI features for low-grade glioma); brain_metastasis; meningioma; schwannoma; lymphoma; pineal_tumor (“yes” for any tumor in the pineal region); intraventricular_tumor; pediatric_posterior_fossa_tumor (diagnosis includes, but not limited to, medulloblastoma, ependymoma, pilocytic astrocytoma); brainstem_glioma (diagnosis also called diffuse midline gliomas); pituitary_tumor; rathkes_cleft_cyst; craniopharyngioma; chordoma_chondrosarcoma; chiari_malformation (“yes” for chiari I/II malformations); cerebral_atrophy; brain_herniation; brain_mass_effect; brain_midline_shift; cephaloceles; encephalomalacia_gliosis; cerebral_edema; cavum_septum_pellucidum; dandy_walker_malformation; mega_cisterna_magna; heterotopia; pachygyria_lissencephaly; schizencephaly; dysgenesis_corpus_callosum; ventriculomegaly (“yes” for any abnormally enlarged ventricles); hydrocephalus_ex_vacuo (“yes” for ventriculomegaly due to brain atrophy, normal pressure hydrocephalus, or ex vacuo dilation); obstructive_hydrocephalus (“yes” for ventriculomegaly due to mass lesion or aqueductal stenosis); subdural_hygroma_effusions; intracranial_hypotension; craniotomy_craniectomy; tumor_resection_cavity; postsurgical_changes; pneumocephalus; catheter (“yes” for any intracranial catheter); spine_syrinx.
hardware and software All data used to train NeuroVFM were acquired over more than two decades of routine clinical care at Michigan Medicine. Studies were identified in the Michigan Medicine PACS via the SDW. The aggregate size of raw NIfTI volumes is ∼150 TB, stored on the ARC DataDen object-storage service.
B.2. SupplementaryData Tables 1. Descriptive characteristics of the UM-NeuroImages prospective test set 2. Full task-level performance on the UM-NeuroImages CT test set 3. Full task-level performance on the UM-NeuroImages MRI test set 4. Performance of NeuroVFM on external neuroimaging benchmarks 5. Generated reports and evaluations for the UM-NeuroImages-Triage test set 6. Hyperparameters for NeuroVFM pretraining and UM-NeuroImages study-level attentive probing 7. Hyperparameters for NeuroVFM-LLaVA training and inference
B.2. Supplementary
SQL Query: Retrieve neuroimaging studies from PACS
📸 Image Gallery