Biologically Inspired LGN-CNN Architecture Mimics Lateral Geniculate Nucleus Functionality
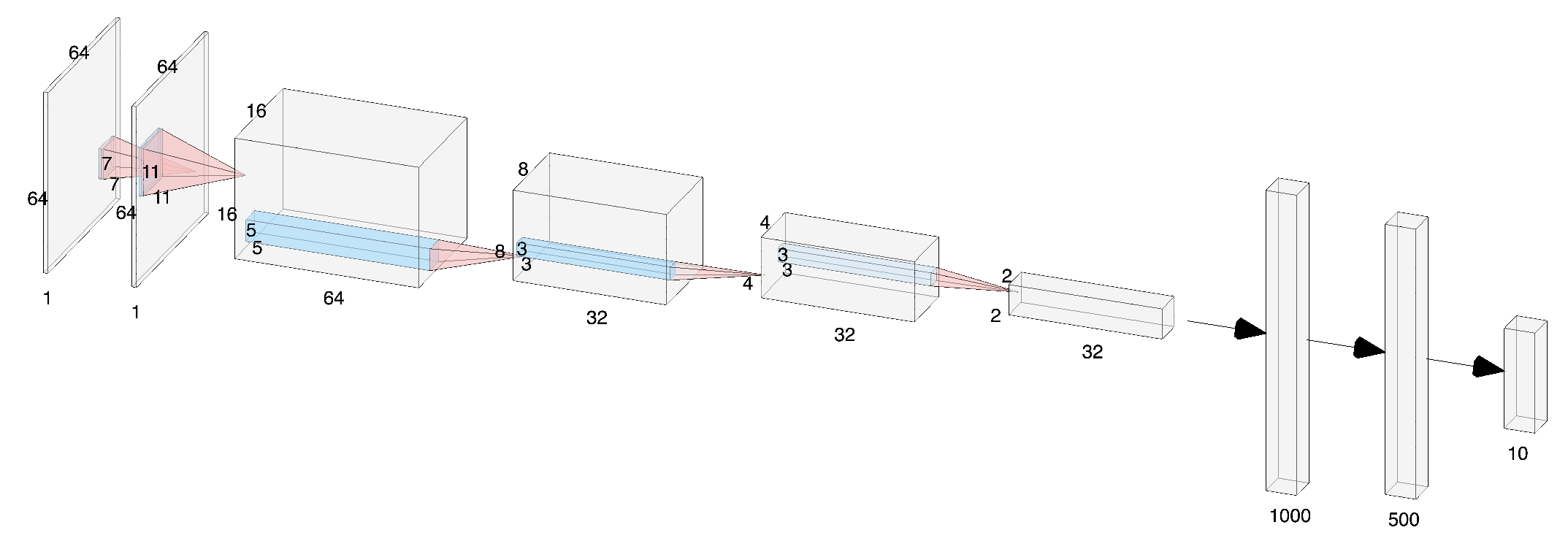
In this paper we introduce a biologically inspired Convolutional Neural Network (CNN) architecture called LGN-CNN that has a first convolutional layer composed by a single filter that mimics the role of the Lateral Geniculate Nucleus (LGN). The first layer of the neural network shows a rotational symmetric pattern justified by the structure of the net itself that turns up to be an approximation of a Laplacian of Gaussian (LoG). The latter function is in turn a good approximation of the receptive field profiles (RFPs) of the cells in the LGN. The analogy with the visual system is established, emerging directly from the architecture of the neural network. A proof of rotation invariance of the first layer is given on a fixed LGN-CNN architecture and the computational results are shown. Thus, contrast invariance capability of the LGN-CNN is investigated and a comparison between the Retinex effects of the first layer of LGN-CNN and the Retinex effects of a LoG is provided on different images. A statistical study is done on the filters of the second convolutional layer with respect to biological data. In conclusion, the model we have introduced approximates well the RFPs of both LGN and V1 attaining similar behavior as regards long range connections of LGN cells that show Retinex effects.