Bayesian Optimization (BO) is a key methodology for accelerating molecular discovery by estimating the mapping from molecules to their properties while seeking the optimal candidate. Typically, BO iteratively updates a probabilistic surrogate model of this mapping and optimizes acquisition functions derived from the model to guide molecule selection. However, its performance is limited in low-data regimes with insufficient prior knowledge and vast candidate spaces. Large language models (LLMs) and chemistry foundation models offer rich priors to enhance BO, but high-dimensional features, costly in-context learning, and the computational burden of deep Bayesian surrogates hinder their full utilization. To address these challenges, we propose a likelihood-free BO method that bypasses explicit surrogate modeling and directly leverages priors from general LLMs and chemistry-specific foundation models to inform acquisition functions. Our method also learns a tree-structured partition of the molecular search space with local acquisition functions, enabling efficient candidate selection via Monte Carlo Tree Search. By further incorporating coarse-grained LLM-based clustering, it substantially improves scalability to large candidate sets by restricting acquisition function evaluations to clusters with statistically higher property values. We show through extensive experiments and ablations that the proposed method substantially improves scalability, robustness, and sample efficiency in LLM-guided BO for molecular discovery.
Discovering molecules with desirable properties is crucial for drug design, materials science, and chemical engineering. Given the vast chemical space (Restrepo, 2022), exhaustive evaluation is infeasible, as density functional theory (DFT) simulations (Parr, 1989) are computationally expensive and experiments are laborious and time-consuming. Bayesian Optimization (BO, Frazier (2018); Garnett (2023)) promises to minimize costly evaluations and accelerate discovery by using acquisition functions, expressed as the expected utility under a surrogate model (e.g., Gaussian Processes (GPs) and Bayesian Neural Networks (BNNs)) to guide the search toward promising candidates, balancing exploration of uncertain regions with exploitation of high observed-value regions.
However, the high cost of evaluations limits the number of initial points available to seed BO with informative Acquisition Function (AF) priors (typically ∼10 in previous studies (Xie et al., 2025;Kristiadi et al., 2024)), further constraining its performance. Recent approaches incorporate LLM priors into BNNs (Kristiadi et al., 2024) with fixed features, parameter-efficient fine-tuning (PEFT; e.g., Low-Rank Adaptation (LoRA (Hu et al., 2021))), or in-context learning (ICL;(Ramos et al., 2023)), but they remain limited by scalability, cost, and computational challenges due to the vast discrete candidate space.
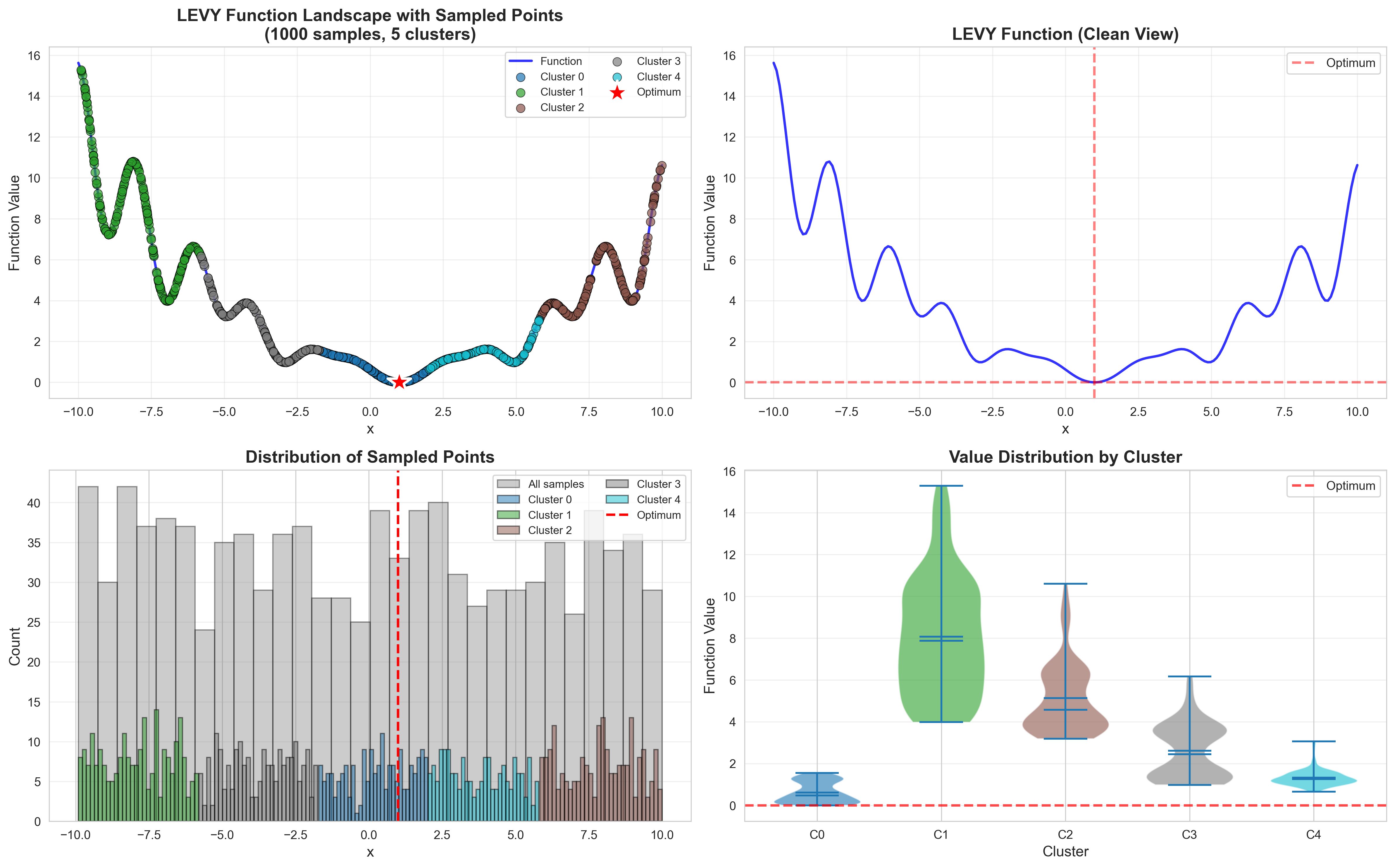
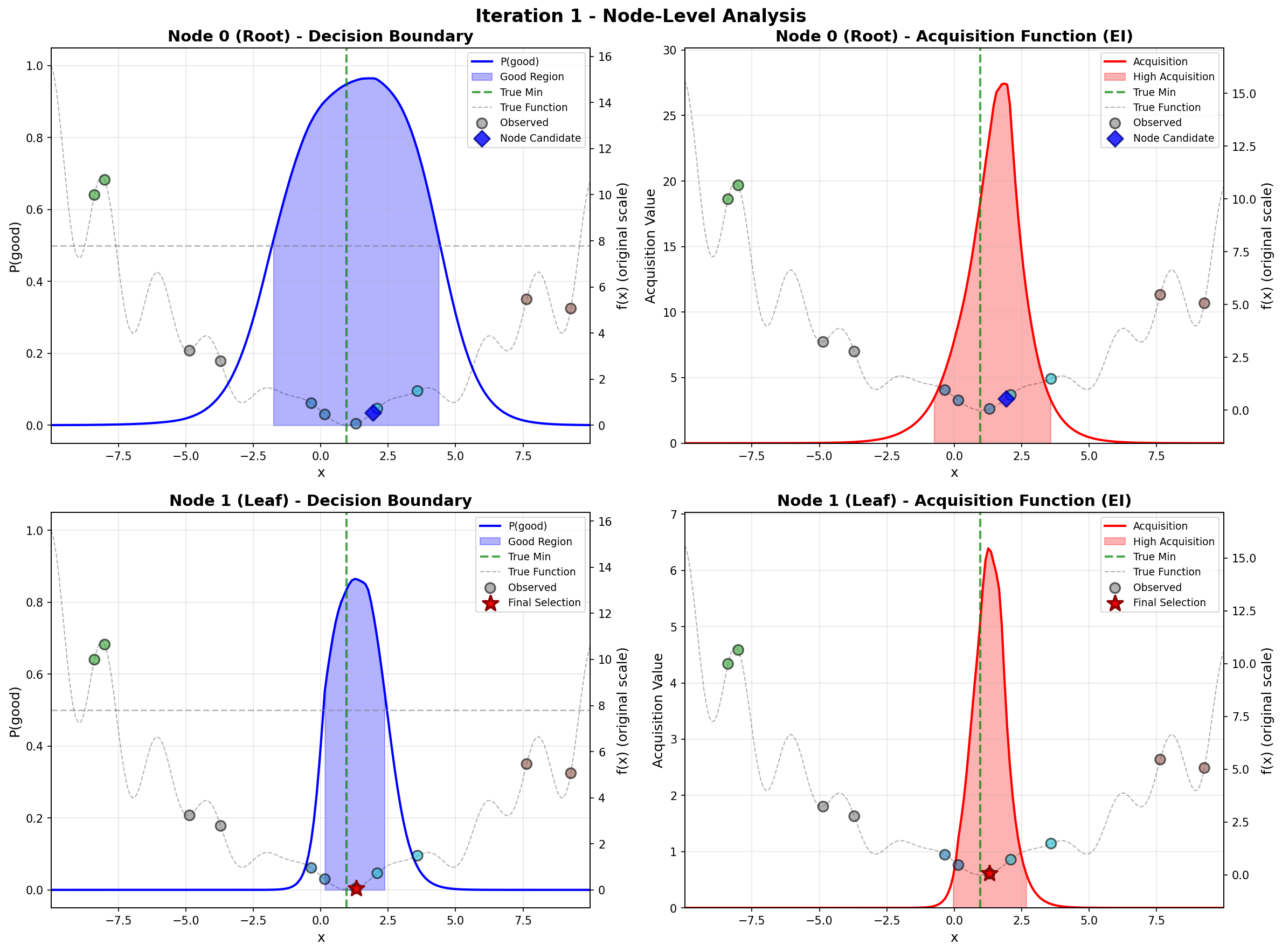
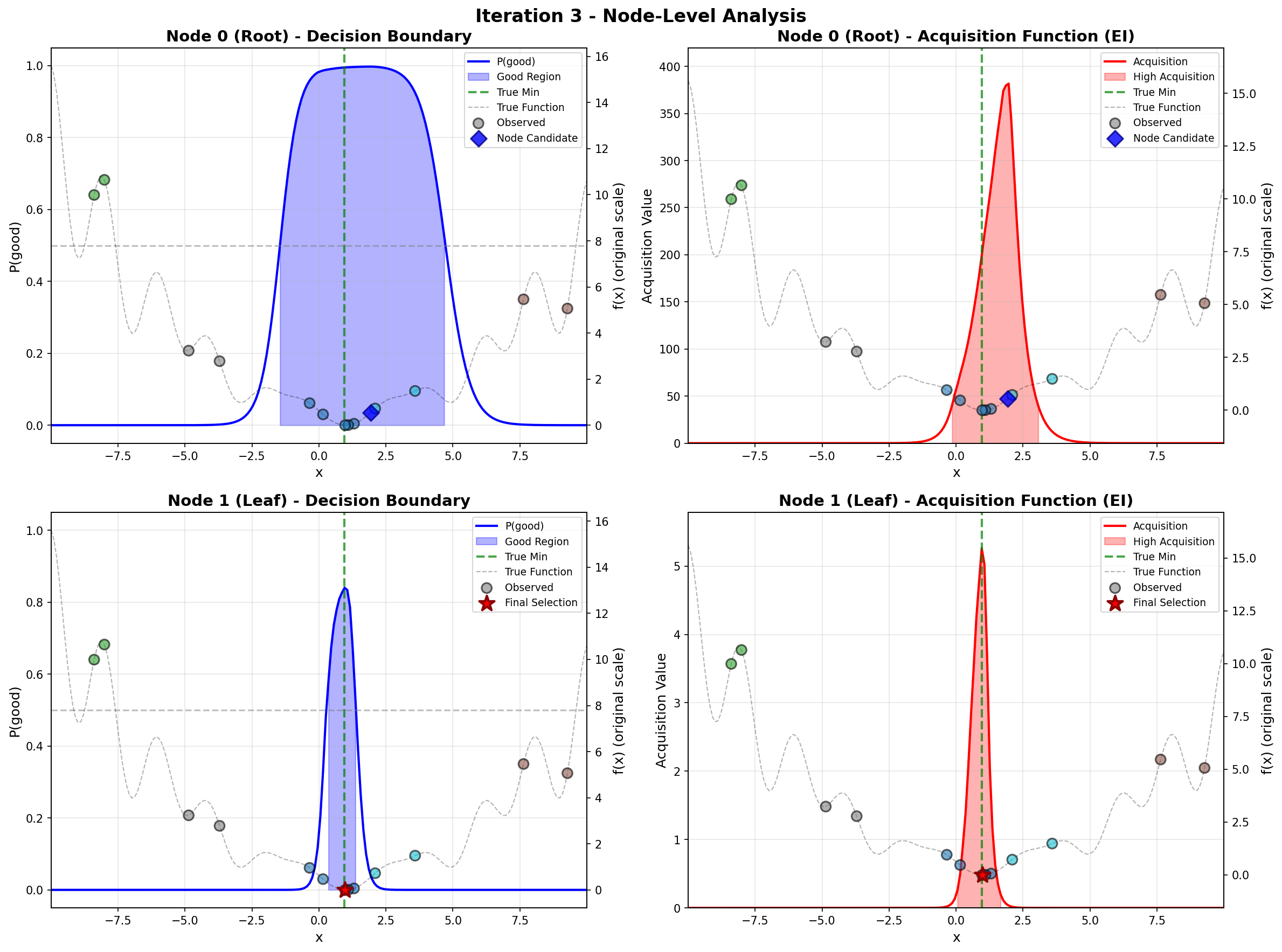
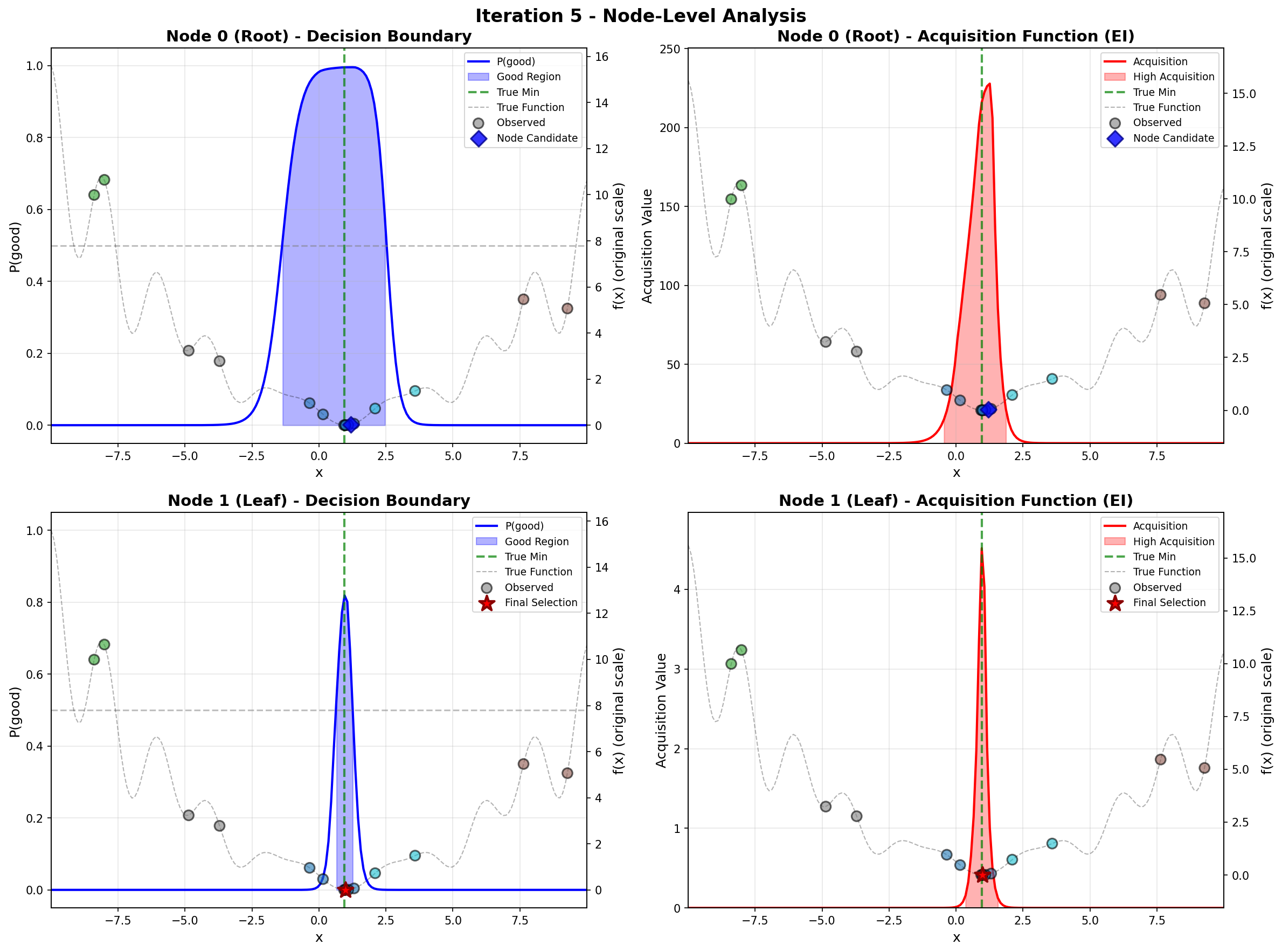
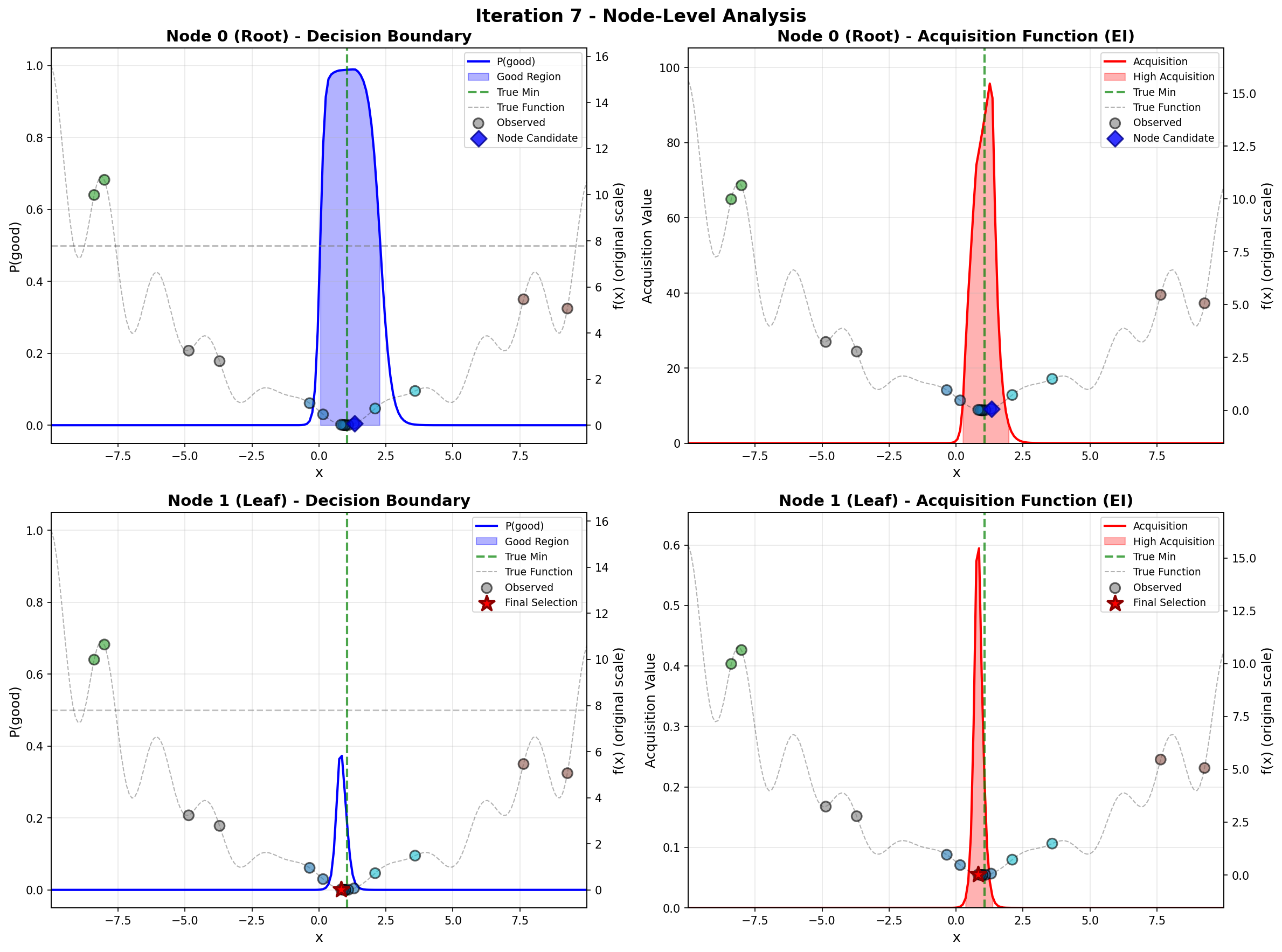
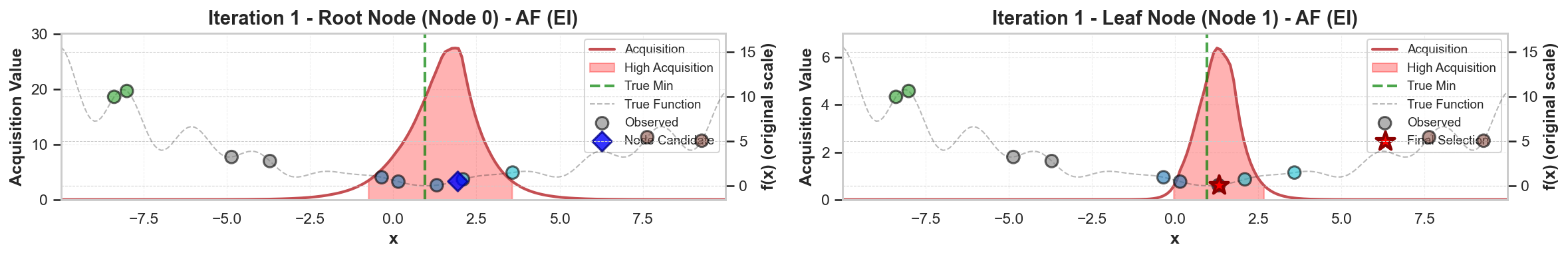
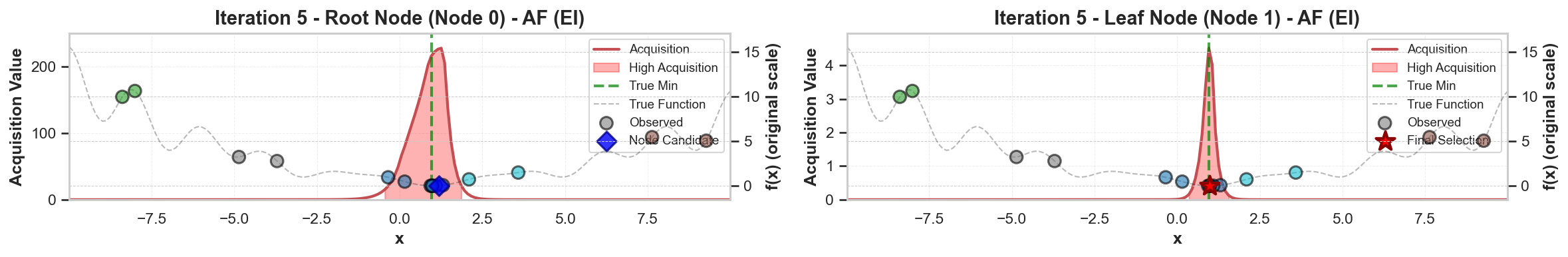
To address these challenges, we propose a principled likelihood-free BO method that avoids the costly Bayesian learning of a surrogate model, which (a) leverages rich prior knowledge from both general LLMs and specialized foundation models to inform AFs, and (b) partitions the molecular candidate space into a tree structure with local AF learned for each node, enabling efficient candidate selection via Monte Carlo Tree Search (MCTS) at each BO iteration given high-dimensional LLM features.
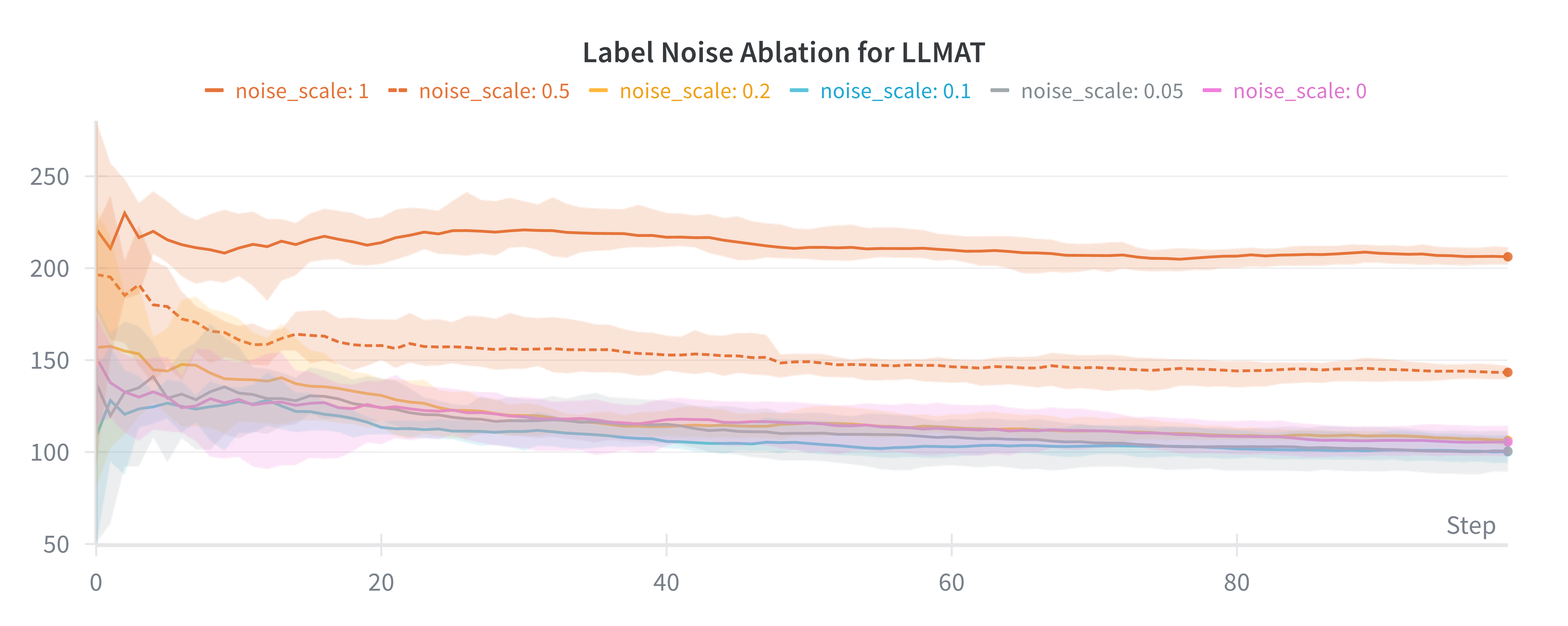
Our method directly models local AFs via density ratio estimation, which can be obtained by optimizing a binary classification objective at each tree node. These binary classifiers determine both the tree partitions and the corresponding local AFs. By meta-learning the shared LoRA weights and the initialization of the root node classifier, we enhance the stability of the binary classifiers, leading to more stable PEFT updates, even in low-data regimes. We further show that our proposed method, LLMAT (LLM-guided Acquisition Tree), visualized in Fig. 2, substantially improves the scalability, robustness, and sample efficiency of BO for molecular discovery. DeepSeek: 2.15 V, 1.42 V, 2.35 V. GPT-4o: 1.42 V, 1.10 V, 2.0 V. Ground Truth: 2.80 V, 1.54 V, 3.38 V.
Prompt: Group {Molecule 1} {Molecule 2} and {Molecule 3} into clusters {0-4, low-high} by redox potential. Respond with a number only. DeepSeek: 3, 1, 4. GPT-4o: 2, 1, 4.
Figure 1: General LLMs provide coarse property ranking of redox potentials rather than accurate numeric value for the three given molecules; responses are color-matched.
Finally, we observed that, although general LLMs are primarily trained on natural language and cannot predict precise numerical property values without in-context learning (ICL), they can capture coarse-grained information, such as whether a molecule’s property is relatively high or low. As shown in Fig. 1, while LLM-predicted redox potentials are not numerically accurate, the relative ordering of molecules is largely preserved. Building on this observation, we introduce an LLM-based clustering phase that queries a general LLM once to assign cluster labels to the entire candidate set. This cluster information is then used to improve scalability and BO performance by restricting AF evaluations to clusters with statistically higher property values.
To summarize, the main contributions of our paper are the following:
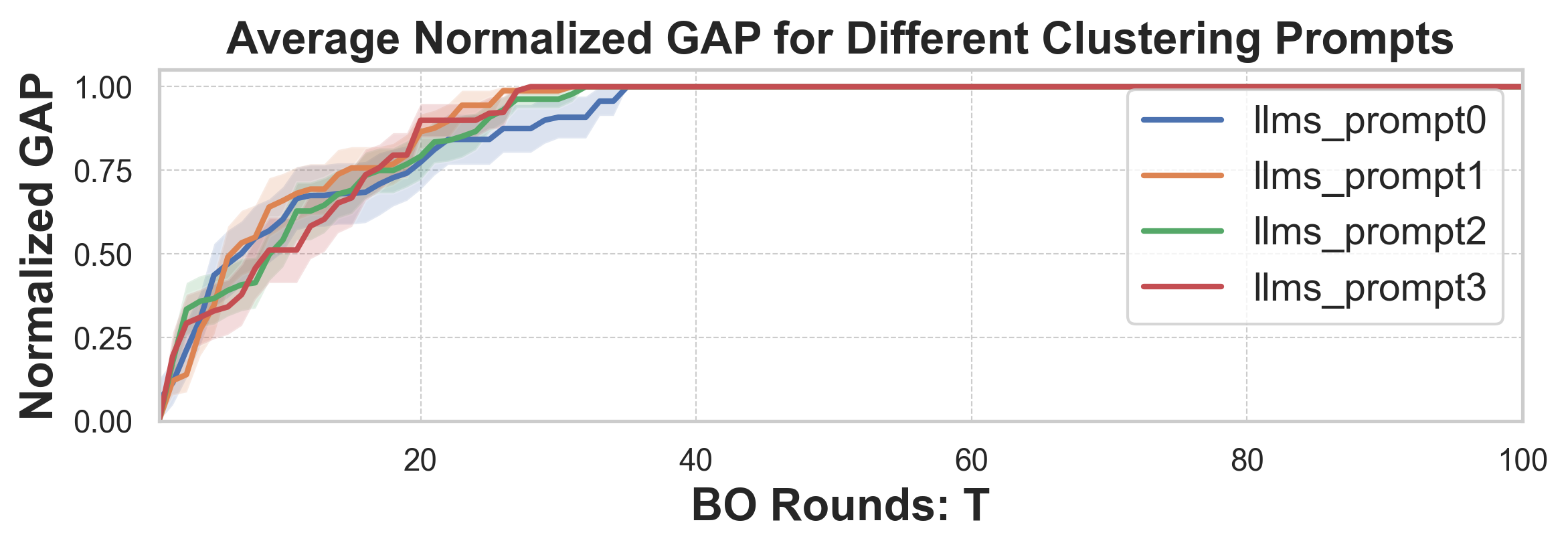
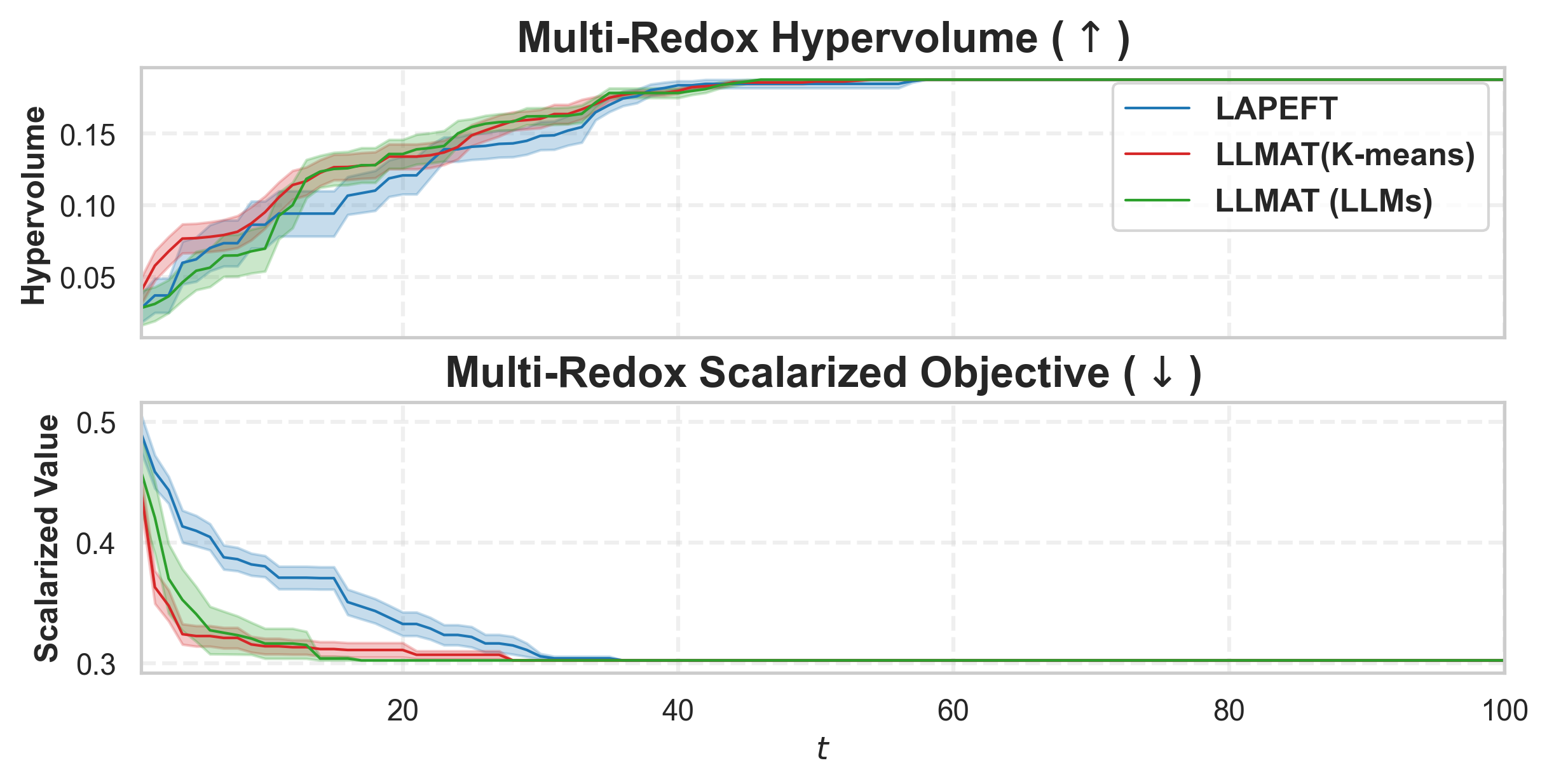
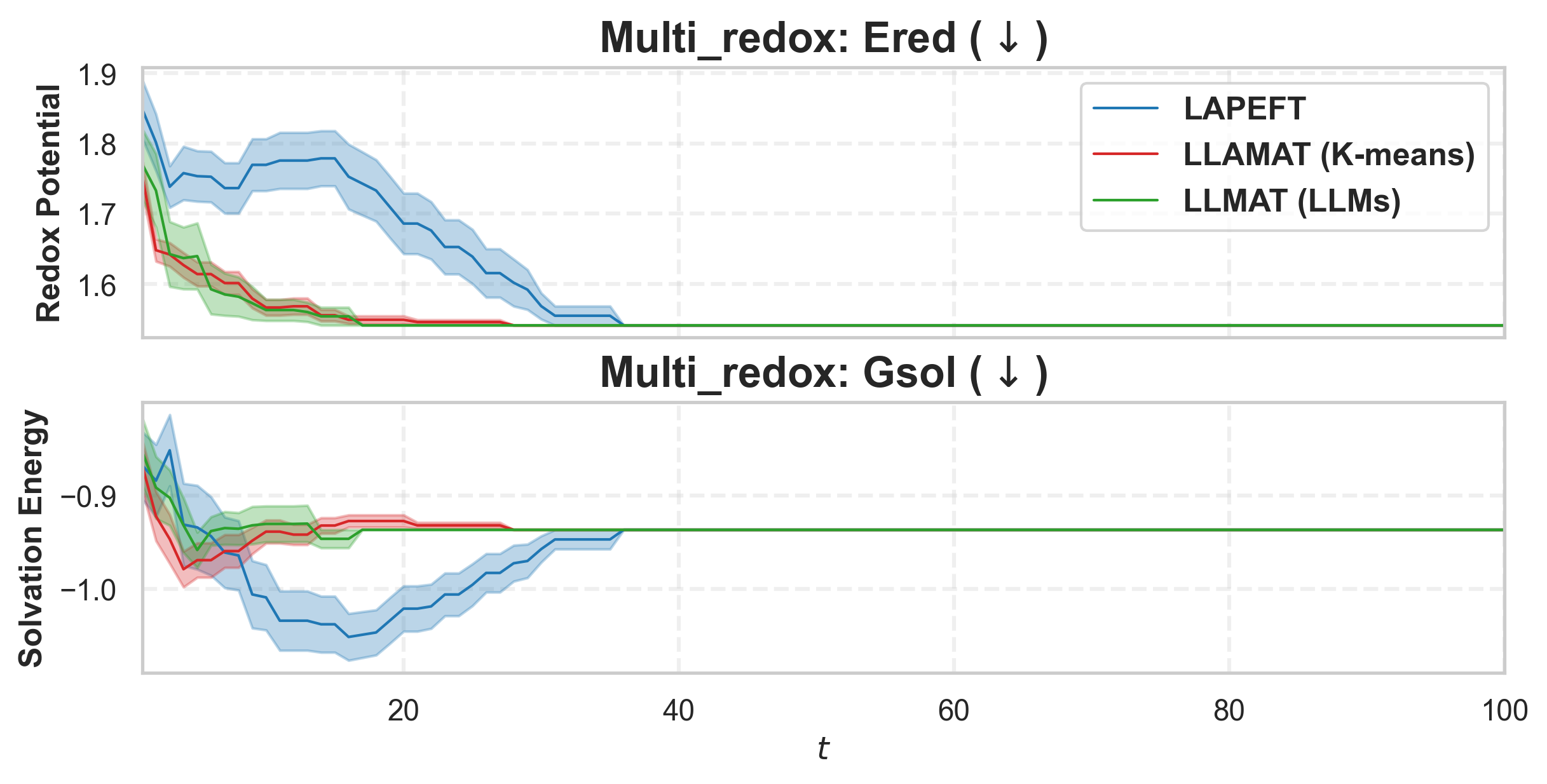
• We propose a likelihood-free BO method that leverages both general LLMs and specialized foundation models to inform AFs in a mathematically principled way. Our method also learns a tree-structured partition of the vast molecular search space and local AFs for the nodes with shared binary classifiers, enabling efficient candidate selection and local AF estimation via MCTS. • To further improve the stability of partitioning and local AFs in the low-data regime, we introduce a meta-learning approach for training the shared binary classifiers, which enables more reliable and generalizable acquisition functions across partitions. • We introduce an LLM-guided pre-clustering strategy along with a statistical cluster selection approach for BO, which estimates AF values only for candidates within selected clusters. This improves BO performance and reduces computational cost, particularly for PEFT, thereby mitigating the scalability challenges associated with vast candidate sets. Our approach provides a novel way to incorporate information from general LLMs into algorithm design with a reasonable API cost.
We evaluate our method on six real-world chemistry datase
This content is AI-processed based on open access ArXiv data.