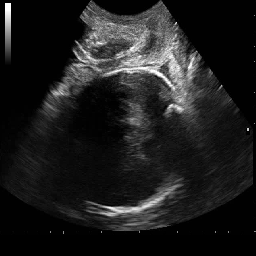
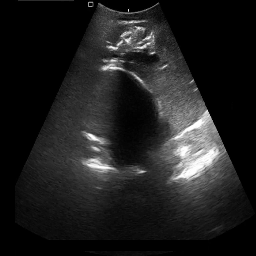
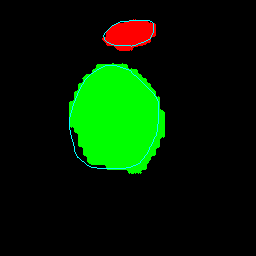
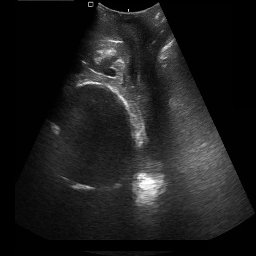
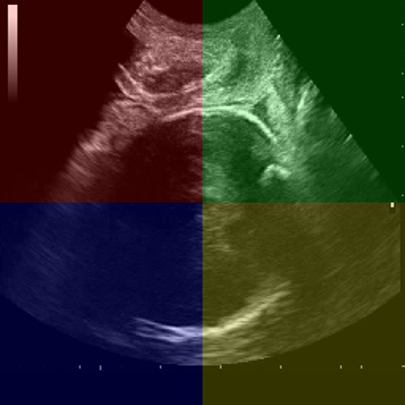
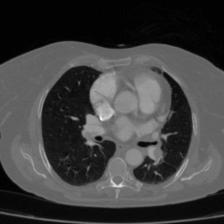
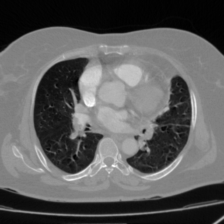
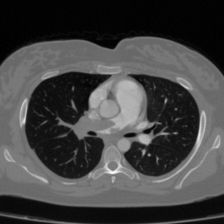
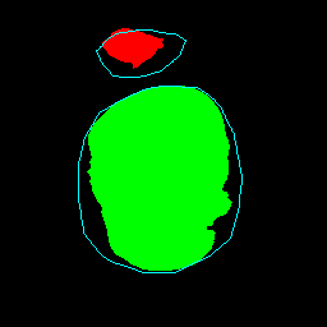
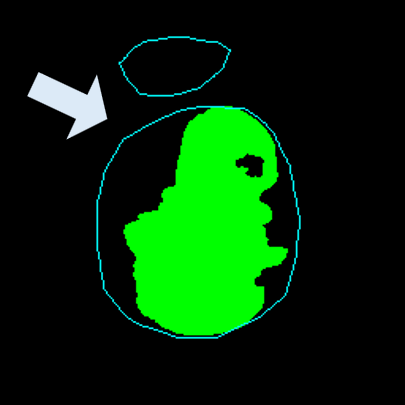
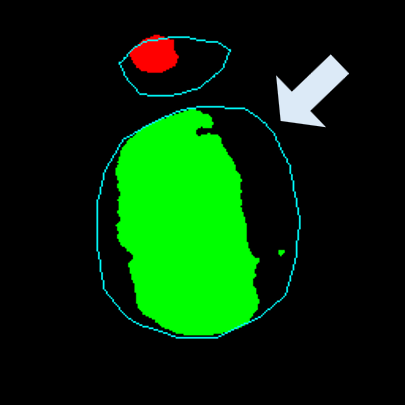
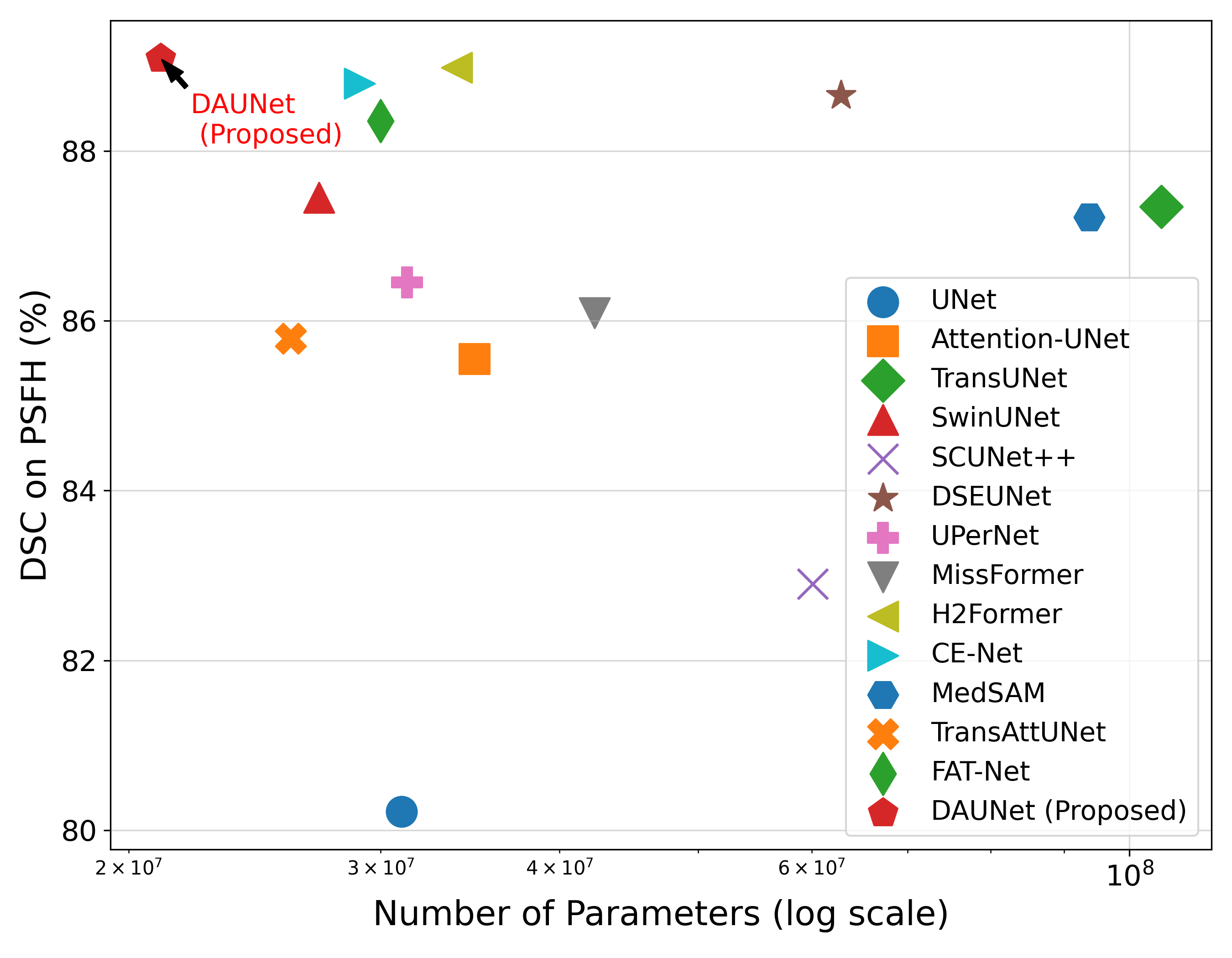
Medical image segmentation plays a pivotal role in automated diagnostic and treatment planning systems. In this work, we present DAUNet, a novel lightweight UNet variant that integrates Deformable V2 Convolutions and Parameter-Free Attention (SimAM) to improve spatial adaptability and context-aware feature fusion without increasing model complexity. DAUNet's bottleneck employs dynamic deformable kernels to handle geometric variations, while the decoder and skip pathways are enhanced using SimAM attention modules for saliency-aware refinement. Extensive evaluations on two challenging datasets, FH-PS-AoP (fetal head and pubic symphysis ultrasound) and FUMPE (CT-based pulmonary embolism detection), demonstrate that DAUNet outperforms state-of-the-art models in Dice score, HD95, and ASD, while maintaining superior parameter efficiency. Ablation studies highlight the individual contributions of deformable convolutions and SimAM attention. DAUNet's robustness to missing context and low-contrast regions establishes its suitability for deployment in real-time and resource-constrained clinical environments.
Medical image segmentation is a foundational task in computer-assisted diagnosis, enabling the precise localization and delineation of anatomical structures that are critical for clinical interpretation, surgical planning, and disease monitoring. Accurate and automated segmentation reduces manual effort and inter-observer variability, particularly in high-throughput clinical settings. Despite significant advances achieved through convolutional neural networks (CNNs), especially the widely adopted UNet architecture [1], key challenges persist, most notably in achieving robustness, han-dling anatomical variability, and maintaining computational efficiency.
Although effective in many scenarios, the classical UNet architecture presents several limitations. Its use of fixedgrid convolutions restricts adaptability to variable-sized features and irregular organ boundaries. Recent deformableconvolution segmentation networks have also emphasized boundary-aware modeling to better align predictions with anatomical contours [2]. Additionally, UNet often struggles in low-contrast or noisy environments, common in modalities such as ultrasound [3], [4] and CT angiography, where anatomical boundaries are not clearly visible [5], [6]. Moreover, UNet lacks mechanisms to capture long-range dependencies, which are crucial for modeling global context in complex medical images.
To overcome these shortcomings, recent works have explored enhancements to UNet via transformer-based modules and attention mechanisms [7], [8]. For instance, Masoudi et al. [7] proposed FAT-Net, which augments a UNet-style backbone with transformer branches to capture long-range interactions and feature adaptation modules to suppress background noise. Similarly, Zhang et al. [9] introduced TransAttUNet, incorporating a Self-Aware Attention (SAA) module that integrates Transformer Self-Attention (TSA) and Global Spatial Attention (GSA) to improve multi-scale feature fusion. Other methods such as DSEUNet [10] and MISSFormer [11] attempt to bridge CNN and transformer paradigms. DSEUNet deepens the UNet backbone while introducing Squeeze-and-Excitation (SE) blocks [12] and hierarchical supervision. MISSFormer, on the other hand, employs enhanced transformer blocks and multi-scale fusion to balance local and global feature representation.
General-purpose models such as MedSAM [13], adapted from the Segment Anything Model (SAM), offer promptbased segmentation across various modalities. Trained on over 1.5 million image-mask pairs, MedSAM shows strong performance on CT, MRI, and endoscopy images. However, limitations remain due to the underrepresentation of certain modalities (e.g., mammography) and imprecise vessel boundary segmentation when using bounding-box prompts.
While the aforementioned models demonstrate commendable segmentation performance, they often suffer from high computational complexity and slower inference, limiting their suitability for real-time or resource-constrained environments.
Hybrid models like H2Former [14] and SCUNet++ [15] further aim to unify the strengths of CNNs and transformers. H2Former leverages hierarchical token-wise and channelwise attention to model both local and global dependencies. SCUNet++ integrates CNN bottlenecks and dense skip connections to improve pulmonary embolism (PE) segmentation. Although SCUNet++ achieves high Dice scores on PE datasets, it tends to produce blocky segmentation outputs on large lesions and has a substantial parameter burden. Other methods, such as CE-Net [16], augment UNet with Dense Atrous Convolution (DAC) and Residual Multi-kernel Pooling (RMP) blocks to improve feature representation, but their multi-branch architectures increase memory requirements and limit scalability.
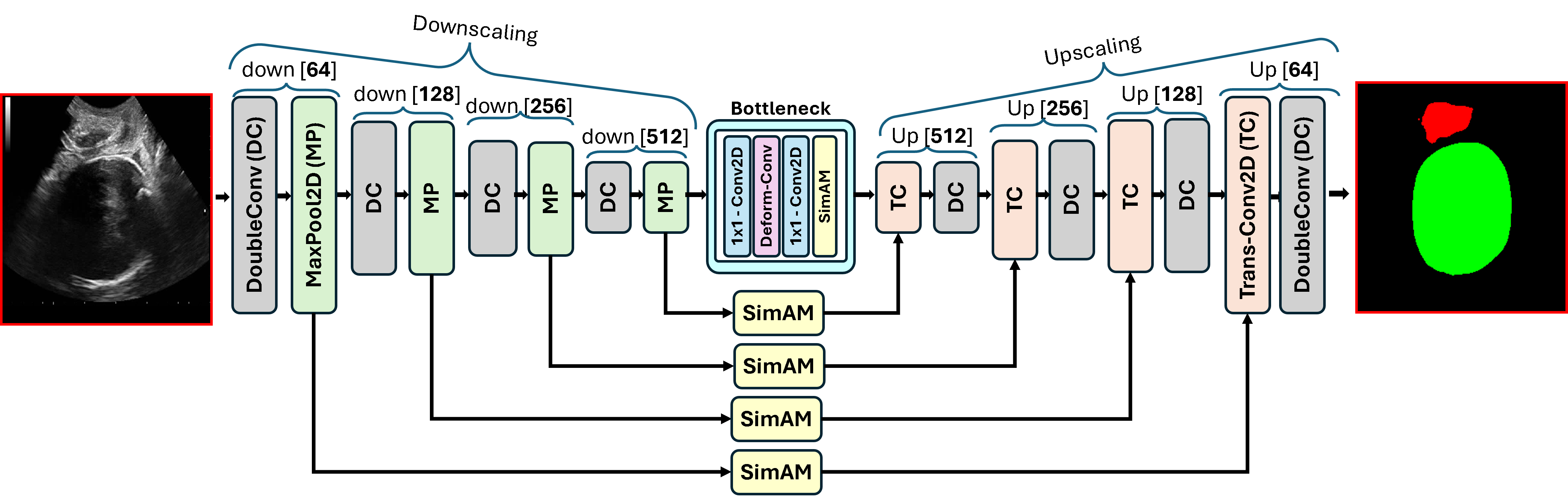
Motivated by the need for efficient, adaptable, and robust segmentation models suitable for real-world clinical deployment, we propose DAUNet, a lightweight and effective UNetbased architecture featuring two key innovations:
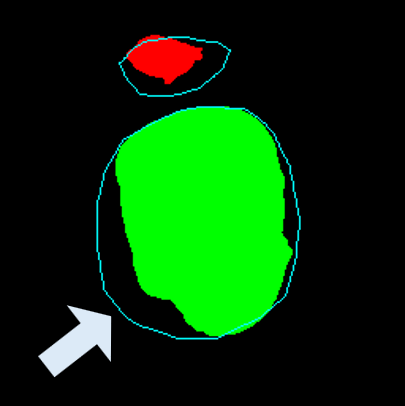
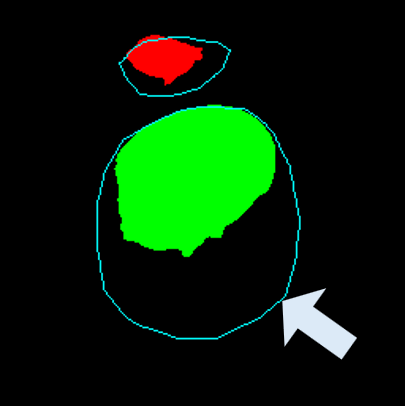
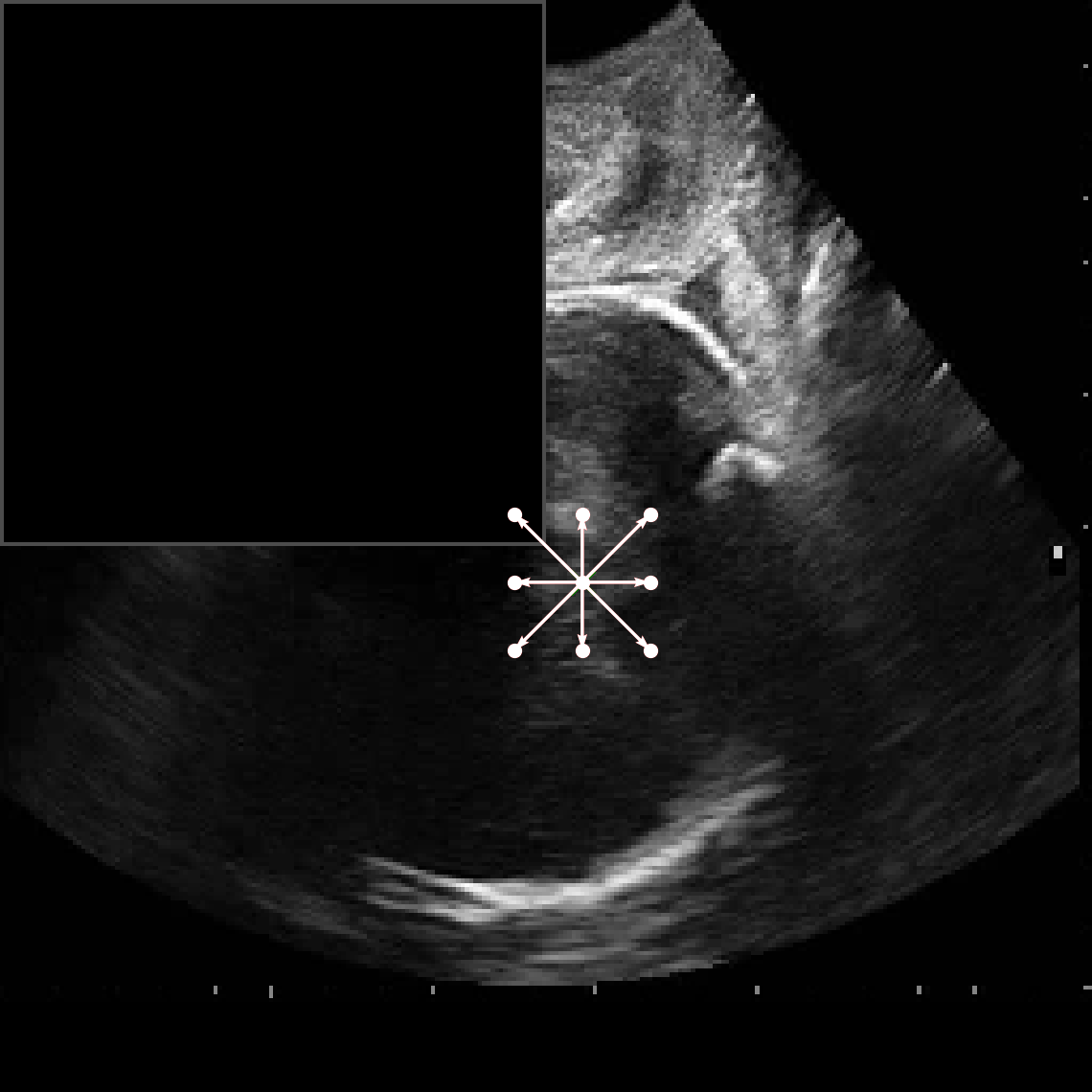
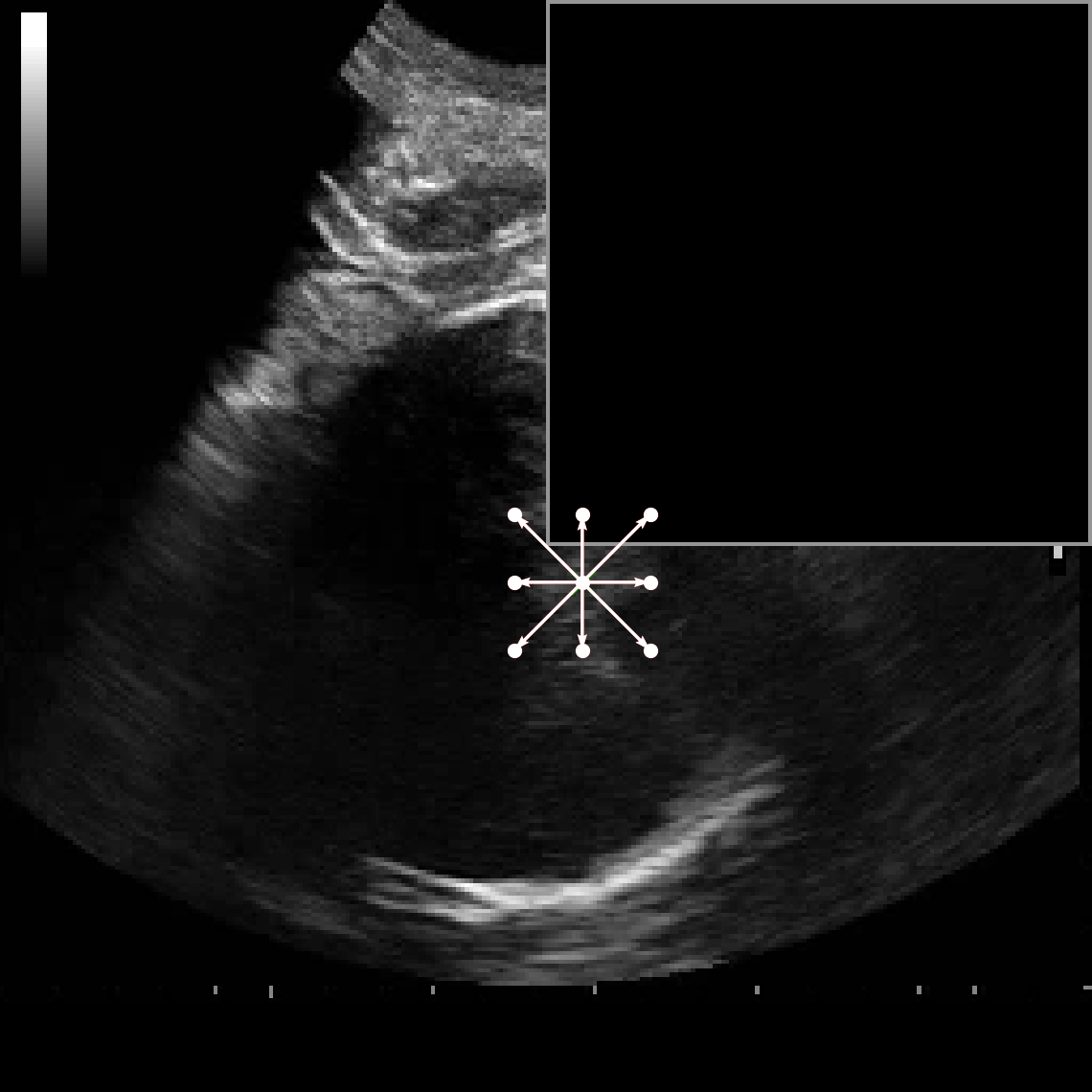
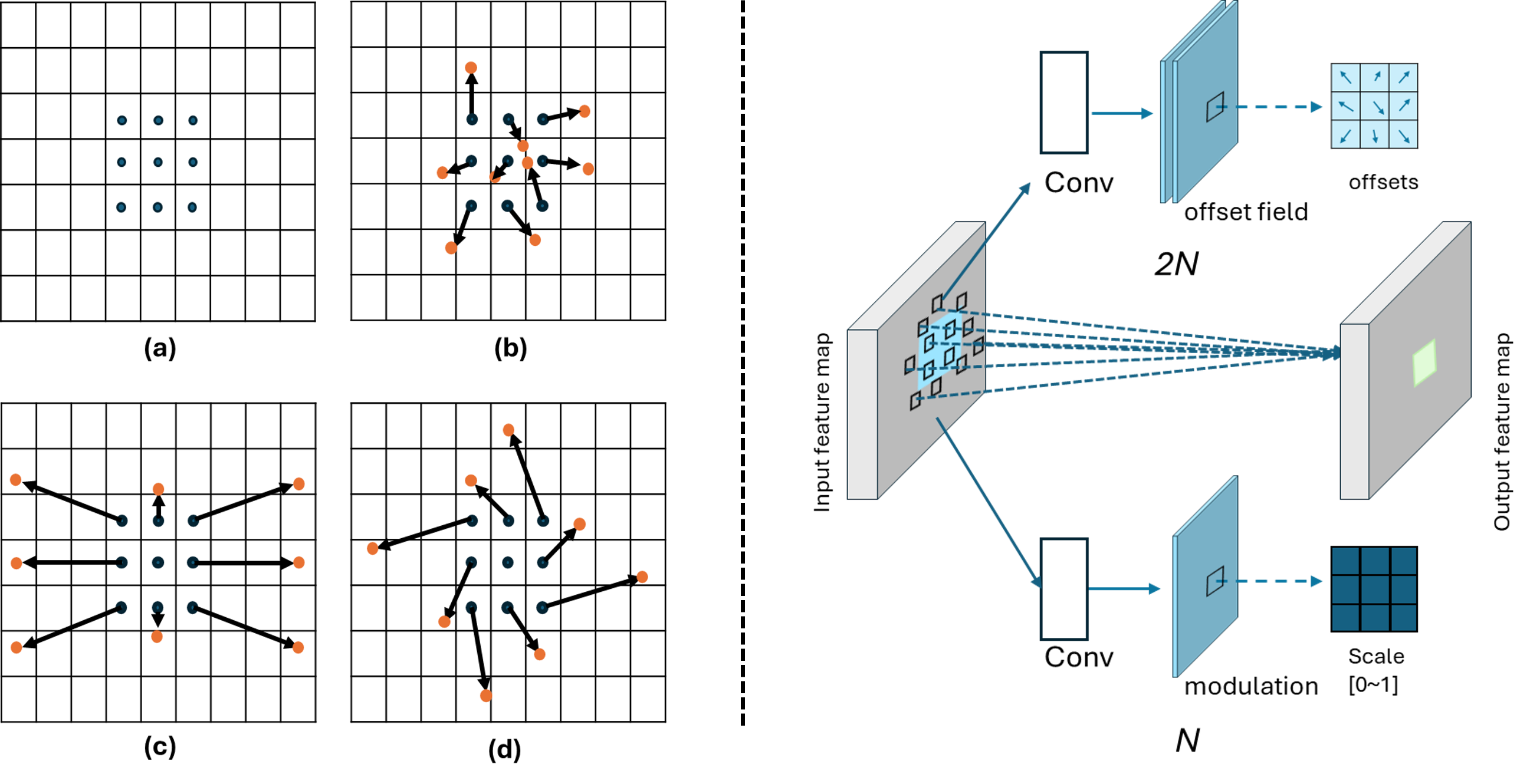
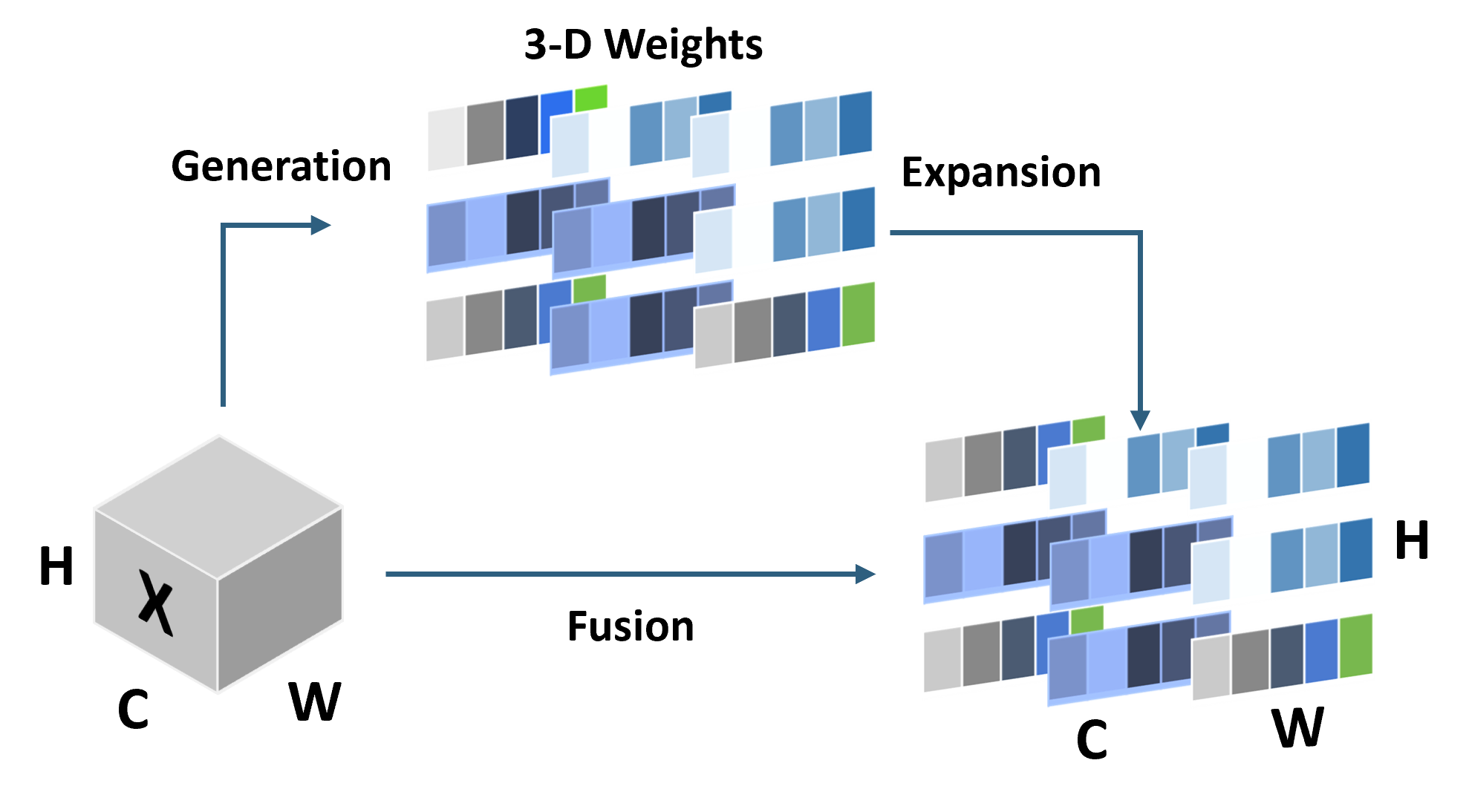
• Improved Bottleneck: A lightweight deformable convolution-based bottleneck module [17], [18] that introduces dynamic, spatially adaptive receptive fields. This design enables the model to better capture geometric deformations and irregular anatomical boundaries. • Improved Decoder: A parameter-free attention mechanism (SimAM) [19] is integrated into the decoder and skips connections to enhance spatial feature representation and facilitate efficient feature fusion, without increasing model complexity. To demonstrate its effectiveness, we evaluate DAUNet on two challenging medical image segmentation tasks: (1) fetal head and pubic symphysis segmentation from transperineal ultrasound using the FH-PS-AoP dataset [20], and (2) pulmonary embolism detection in CT angiography using the FUMPE dataset [21]. Both tasks are characterized by substantial anatomical variability, low-contrast regions, and limited contextual information, factors that commonly impair the performance of conventional models. T
This content is AI-processed based on open access ArXiv data.