Deep learning models have shown high accuracy in classifying electrocardiograms (ECGs), but their black box nature hinders clinical adoption due to a lack of trust and interpretability. To address this, we propose a novel three-stage training paradigm that transfers knowledge from multimodal clinical data (laboratory exams, vitals, biometrics) into a powerful, yet unimodal, ECG encoder. We employ a self-supervised, joint-embedding pre-training stage to create an ECG representation that is enriched with contextual clinical information, while only requiring the ECG signal at inference time. Furthermore, as an indirect way to explain the model's output we train it to also predict associated laboratory abnormalities directly from the ECG embedding. Evaluated on the MIMIC-IV-ECG dataset, our model outperforms a standard signal-only baseline in multi-label diagnosis classification and successfully bridges a substantial portion of the performance gap to a fully multimodal model that requires all data at inference. Our work demonstrates a practical and effective method for creating more accurate and trustworthy ECG classification models. By converting abstract predictions into physiologically grounded \emph{explanations}, our approach offers a promising path toward the safer integration of AI into clinical workflows.
Electrocardiogram (ECG) exams are a fundamental diagnostic tool in medical practice, recording the heart's electrical activity to detect a wide range of cardiovascular conditions [Braunwald et al., 2015]. Their importance is substantial, especially considering that cardiovascular diseases remain the leading cause of global mortality [Roth et al., 2020]. With the advancement of artificial intelligence, the automatic classification of ECG exams using deep learning techniques has shown remarkable potential [Liu et al., 2021, Petmezas et al., 2022], offering high precision in identifying abnormalities and extracting complex information that can aid in diagnosis.
Despite the promising performance and significant accuracy achieved by deep learning models in healthcare, their adoption and trust among healthcare professionals remain a challenge [Reyes et al., 2020, Adeniran et al., 2024]. The main barrier to integrating these models into clinical practice lies in their black box nature-the lack of explainability and transparency about how decisions are made [Rosenbacke et al., 2024]. Physicians, accustomed to clinical reasoning based on evidence and causality, hesitate to trust systems whose internal processes are opaque, leading to mistrust and uncertainty regarding the safety and justification of the predictions [Koçak et al., 2025].
Frequently, explanations are presented as saliency maps, which often fail to bridge the trust gap due to a lack of robustness and misalignment with clinical concepts [Borys et al., 2023, Zhang et al., 2023]. To address this, we propose a novel multimodal training architecture that enriches an ECG model with knowledge from associated tabular clinical data. Instead of requiring all data modalities at inference, our approach uses a self-supervised, joint-embedding objective to transfer the rich context from laboratory values and vitals into a powerful, unimodal ECG encoder. By training it to perform a secondary task of predicting lab abnormalities from the ECG signal alone, we create a system that can explain its diagnostic reasoning in terms of concrete, clinically relevant concepts.
In summary, we make the following contribution:
• We propose a joint-embedding pre-training framework to transfer knowledge from multimodal tabular data into a unimodal ECG encoder for the task of diagnosis classification; • We introduce the prediction of laboratory abnormalities from the ECG embedding as a novel, clinically-grounded method for a indirect explanation of the model’s diagnostic outputs.
Our work integrates insights from three key areas. While deep learning for ECG classification is well-established, models often lack the trust of clinicians due to their “black box” nature [Reyes et al., 2020]. Current eXplainable AI (XAI) methods often rely on saliency maps, which can be misaligned with clinical reasoning [Borys et al., 2023]. Concurrently, self-supervised learning (SSL) has enabled the creation of powerful representations without labeled data [Zbontar et al., 2021], and multimodal learning seeks to create holistic models by fusing data sources [Kline et al., 2022]. However, most multimodal approaches require all data at inference time, hindering practicality [Alcaraz et al., 2025]. We are the first to unify these areas, using a multimodal SSL objective to distill knowledge into a practical, unimodal-at-inference model with a novel, clinically-grounded explanation mechanism. A detailed literature review is provided in the Appendix.
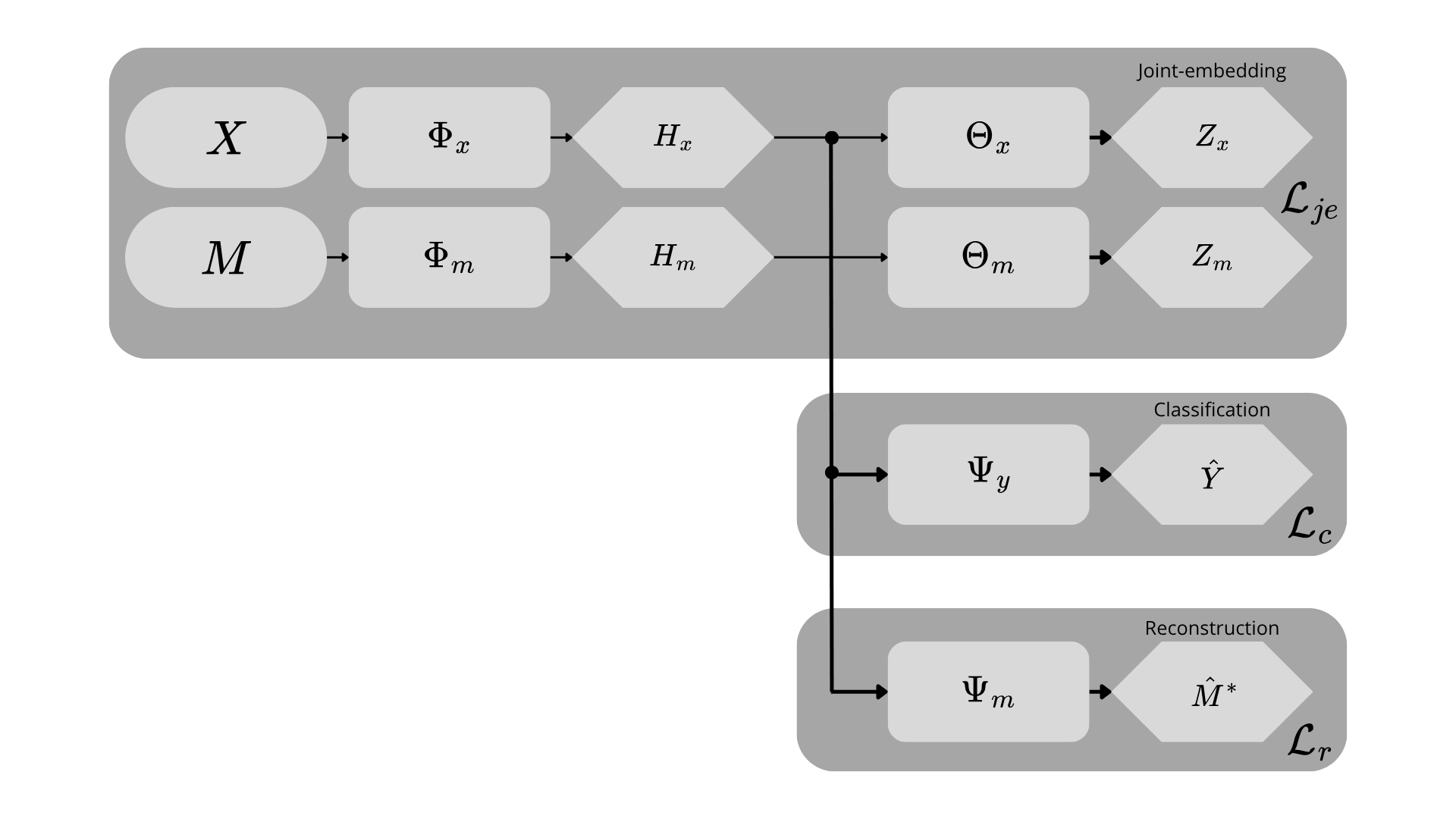
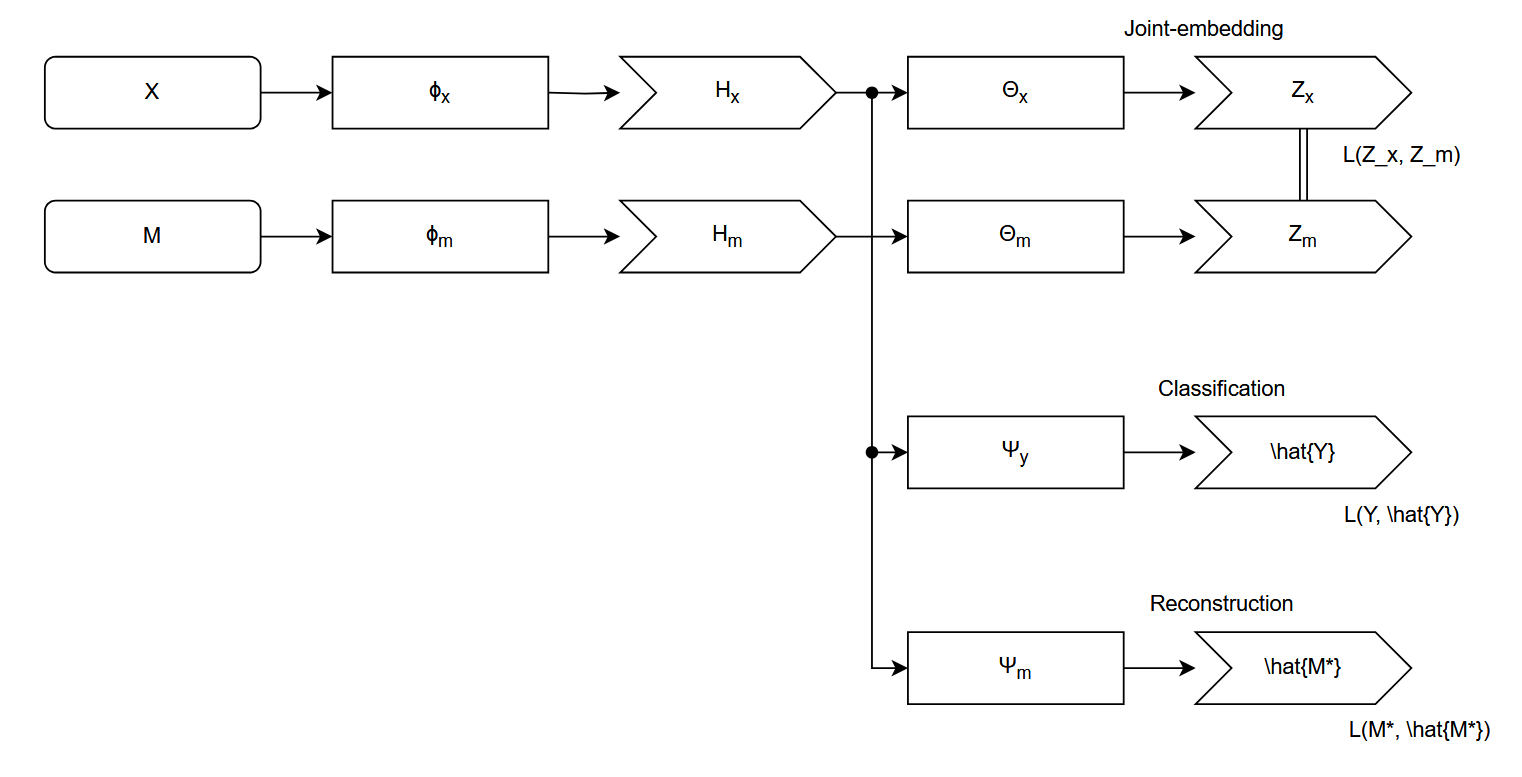
Our approach is a three-stage training paradigm designed to create a powerful ECG representation aligned with clinically relevant outputs. We first pre-train a waveform encoder using a self-supervised, cross-modal objective to transfer knowledge from tabular clinical data. Subsequently, we fine-tune this encoder for two downstream relevant clinical tasks: a primary task of multi-label diagnosis classification Strodthoff et al. [2024]; and, a secondary task of laboratory values abnormality prediction Alcaraz and Strodthoff [2024]. We organised this algorithm into a fluxogram, as despicted in Figure 1.
The primary goal of our pre-training stage is to learn a signal encoder, Φ x , whose representations are enriched with the contextual information present in associated tabular clinical data. To achieve this, we frame the task as a self-supervised, joint-embedding problem where the model learns to align representations from these two distinct modalities.
For a given patient encounter, we have a raw ECG signal segment x ∈ R C×L , where C is the number of leads and L is the sequence length, and a corresponding vector of tabular clinical data m ∈ R D , which includes demographics, vitals, and biometrics. The signal and tabular data are processed by their respective encoders, a powerful S4-based sequence model Φ x [Strodthoff et al., 2024] and a MLP Φ m . These backbones produce feature representations h x = Φ x (x) and h m = Φ m (m).
Following the standard practi
This content is AI-processed based on open access ArXiv data.