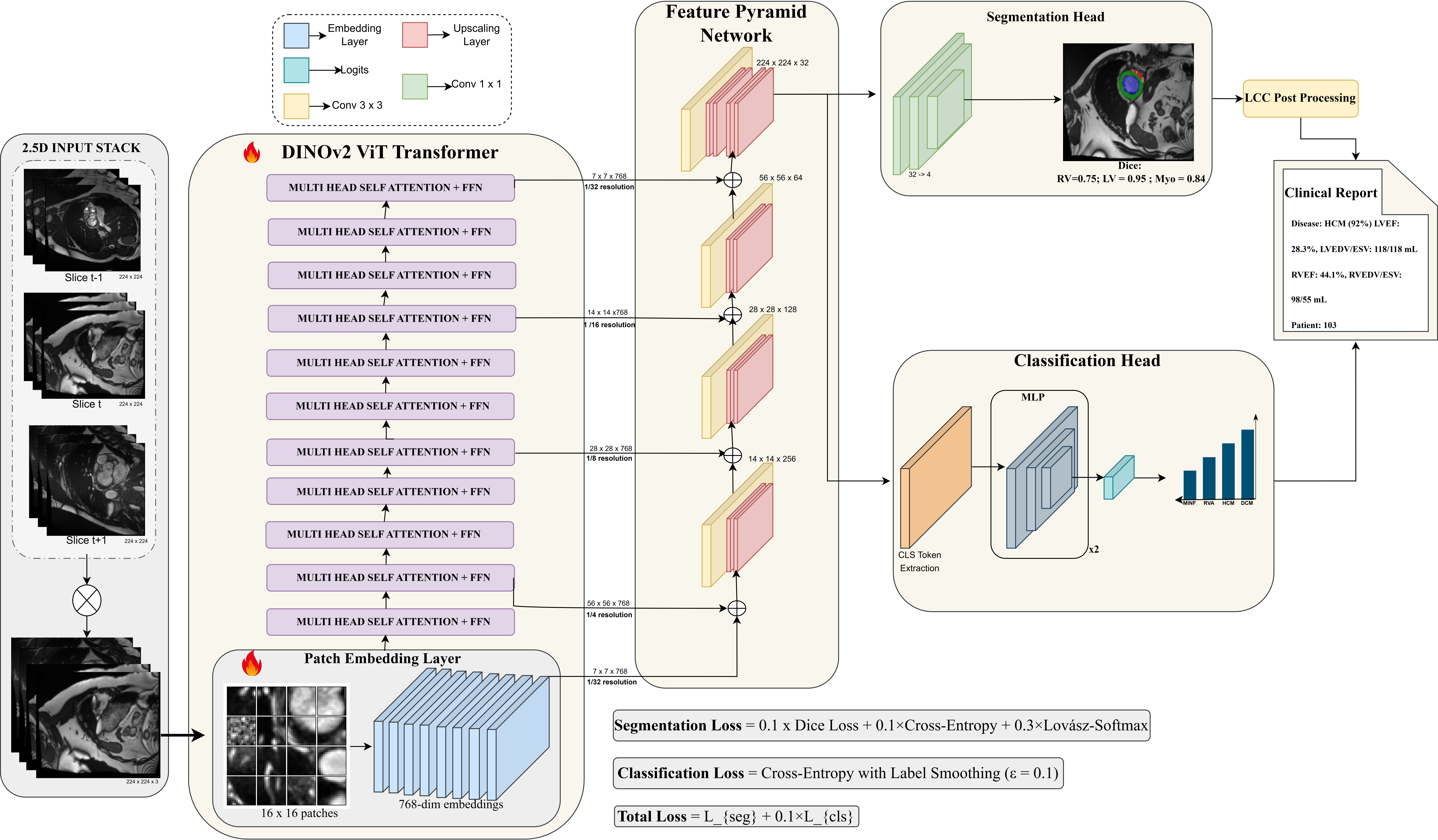
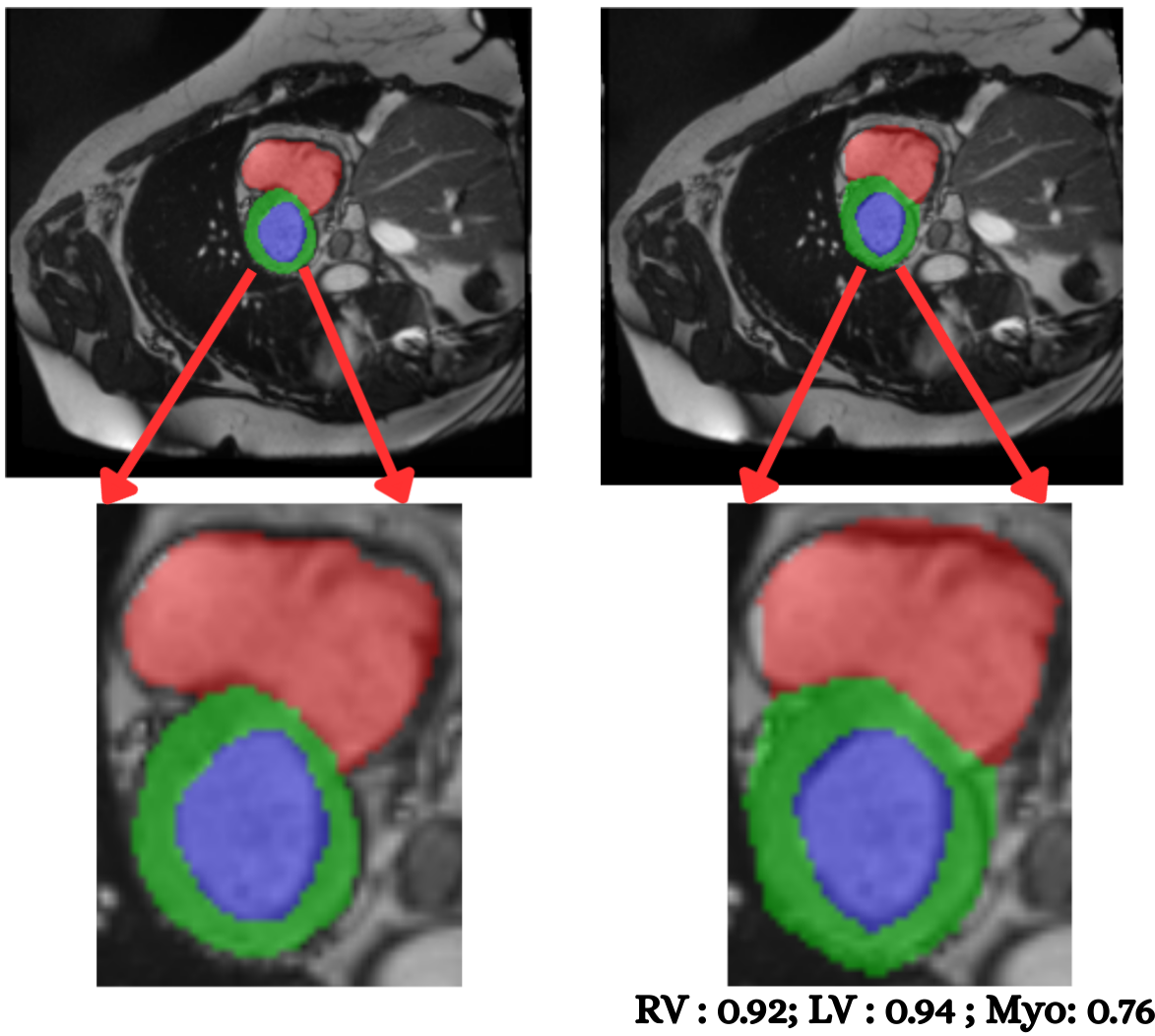
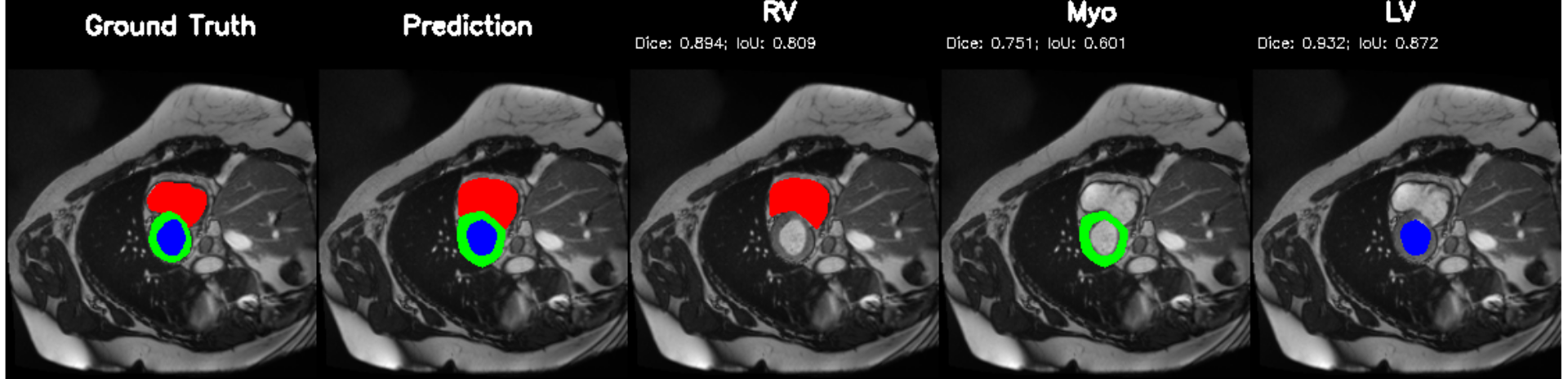
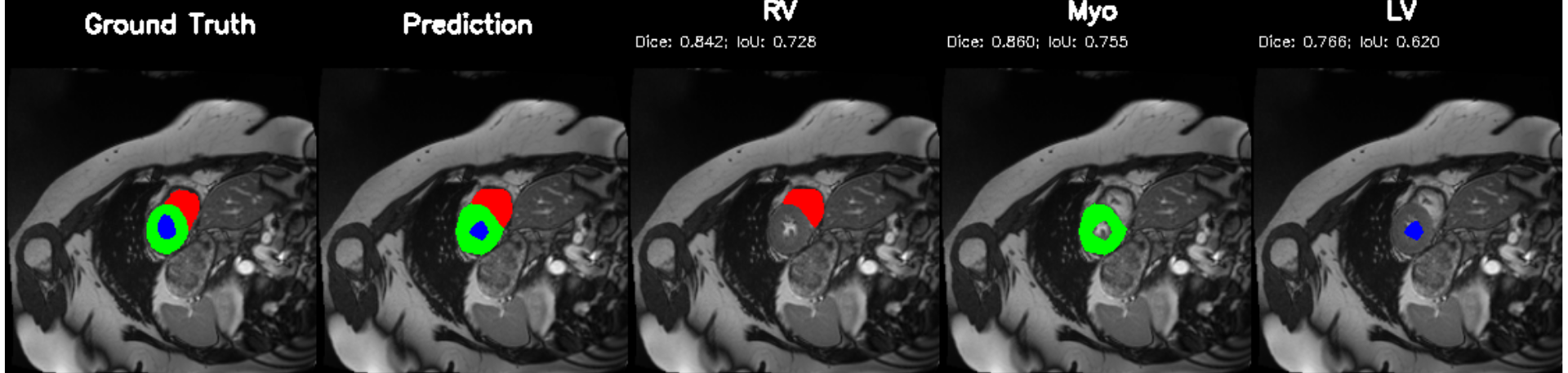
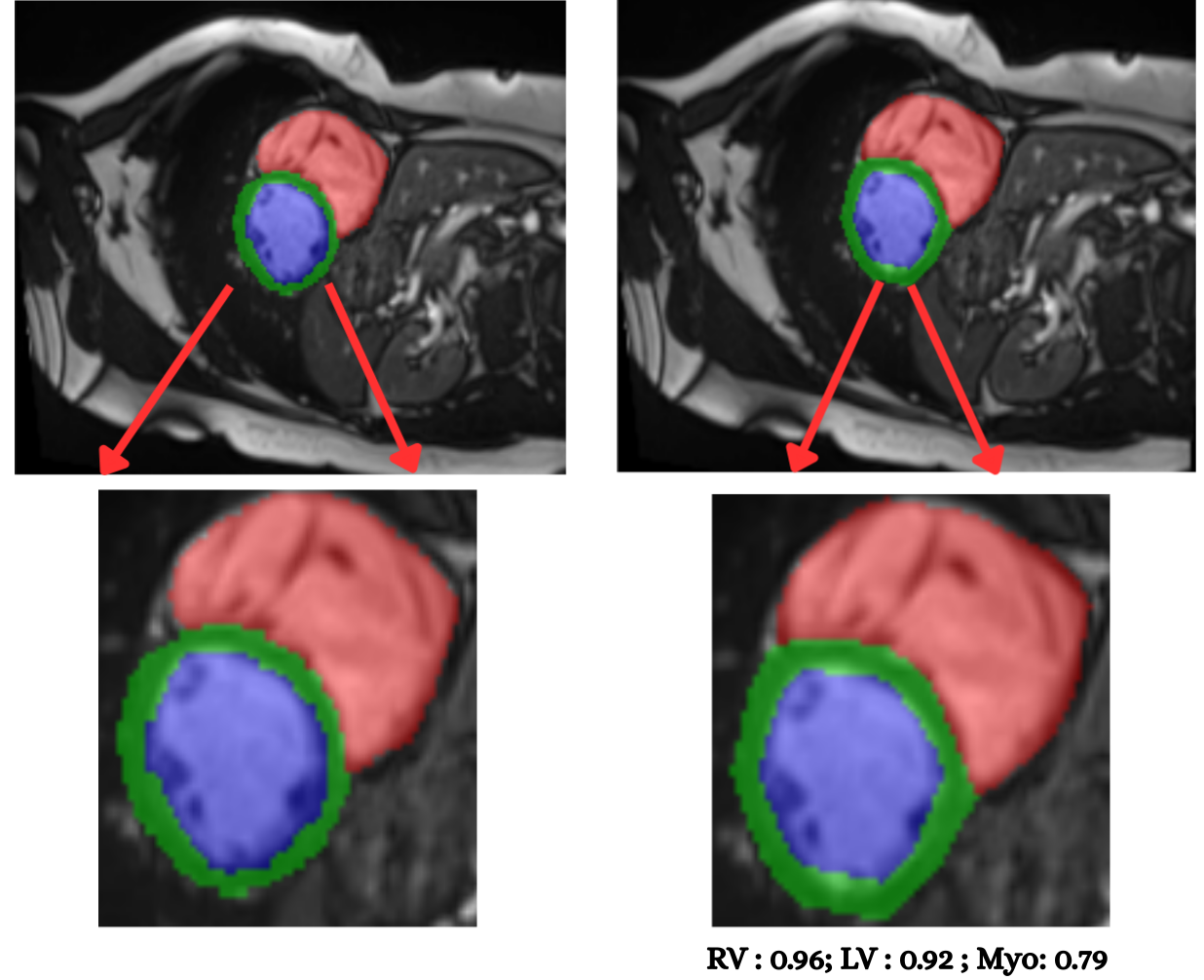
Cardiac image analysis remains fragmented across tasks: anatomical segmentation, disease classification, and grounded clinical report generation are typically handled by separate networks trained under different data regimes. No existing framework unifies these objectives within a single architecture while retaining generalization across imaging modalities and datasets. We introduce PULSE, a multi-task vision-language framework built on self-supervised representations and optimized through a composite supervision strategy that balances region overlap learning, pixel wise classification fidelity, and boundary aware IoU refinement. A multi-scale token reconstruction decoder enables anatomical segmentation, while shared global representations support disease classification and clinically grounded text output allowing the model to transition from pixels to structures and finally clinical reasoning within one architecture. Unlike prior task-specific pipelines, PULSE learns task-invariant cardiac priors, generalizes robustly across datasets, and can be adapted to new imaging modalities with minimal supervision. This moves the field closer to a scalable, foundation style cardiac analysis framework.
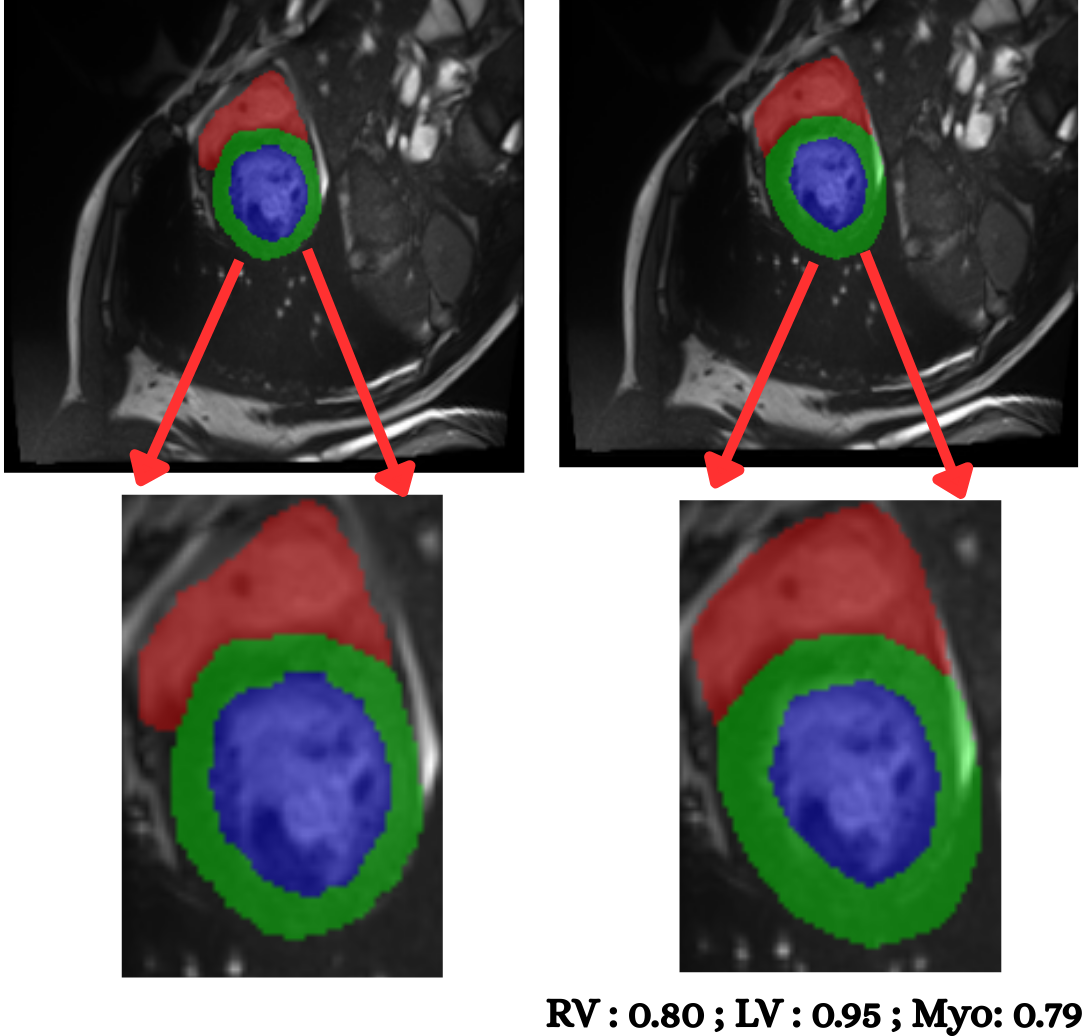
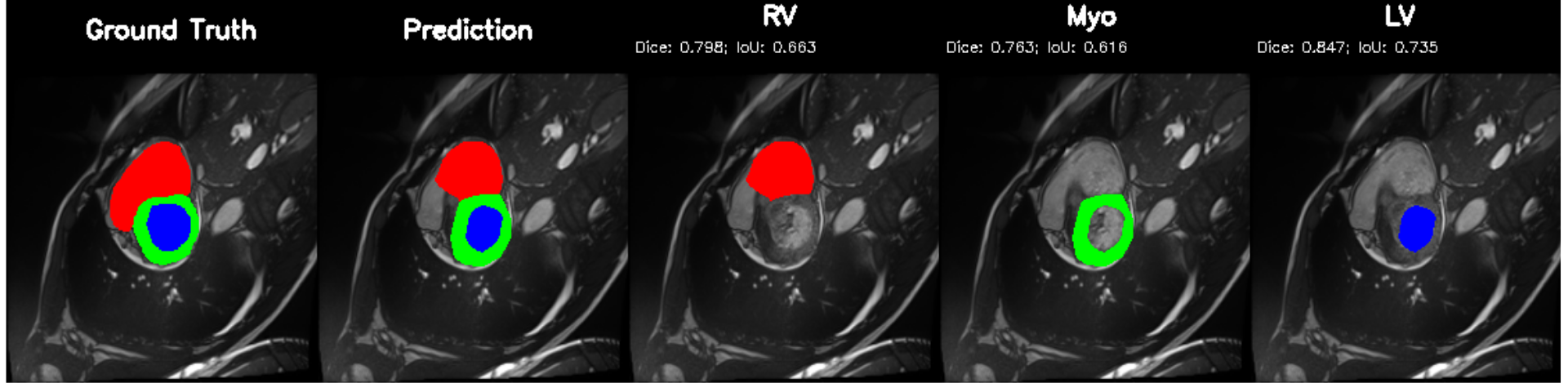
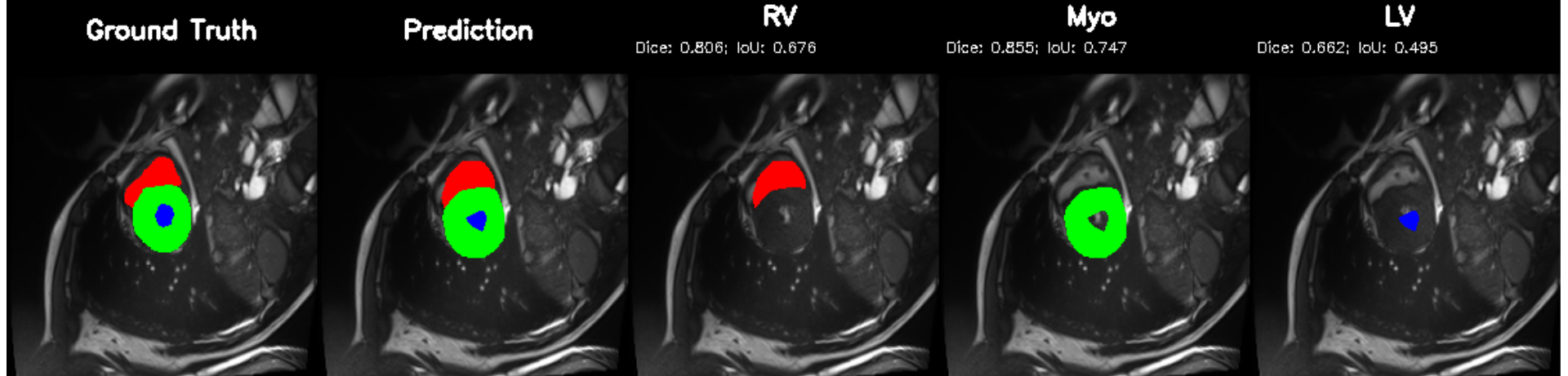
Cardiovascular disease remains the leading cause of mortality worldwide [1]. A core part of its clinical evaluation rests on accurate assessment of the left ventricle (LV), the heart's main pumping chamber, because LV volumes, myocardial mass, and ejection fraction (EF) are fundamental indicators of pump function, remodeling, and disease severity [2,3]. Cardiac magnetic resonance (CMR) imaging is the clinical standard for quantifying ventricular volumes and myocardial thickness, thanks to its superior softtissue contrast and reproducibility [4]; yet segmentation still relies heavily on manual contouring, which is time-consuming, operator-dependent, and hard to scale in high-volume settings [5]. Deep learning (DL) has emerged as the dominant approach for automating cardiac image segmentation, outperforming traditional handcrafted or atlas-based techniques across MRI, CT, and ultrasound modalities [6]. Both Convolutional Neural Networks(CNNs) and newer transformer based architectures now achieve high Dice on benchmark datasets such as the ACDC challenge, reliably delineating ventricular structures even in pathological cases [7,8]. Many pipelines then compute volumetric and functional indices from these masks. However, segmentation alone is insufficient for clinical decision support. Most DL models are developed for a single dataset, typically short-axis MRI cine stacks, under full supervision and do not generalize well across centers, scanner types, vendors, ormodalities, limiting clinical scalability [9].
Independent research efforts performing cardiac disease classification demonstrate that while diagnostic accuracy can be high, they often rely on reduced representations (e.g., global latent vectors, volumes) and discard pixel-wise anatomy provided by segmentation masks [10]. As a result, anatomical segmentation and disease reasoning remain decoupled: one model “sees” structure, another “sees” disease, and the connection between them is mediated only by ad-hoc feature engineering. Real-world clinical workflows demand more: clinicians want a coherent chain from anatomy to quantitative indices to diagnosis and finally to structured report. Yet to our best knowledge, no widely adopted DL framework simultaneously delivers (a) high-fidelity segmentation, (b) disease or functional classification, and (c) clinically relevant outputs (volumetric indices or structured narrative) in a unified pipeline. Even recent cardiac segmentation studies typically conclude at mask generation [8]. Although some multi-task networks attempt to combine segmentation with classification or motion estimation [11], they remain limited: most implement only two tasks (e.g. segmentation + classification), rely on CNN based backbones with limited long range context, and do not produce quantitative indices or structured clinical output . From an architectural perspective, the segmentation only paradigm also overlooks important trade-offs.
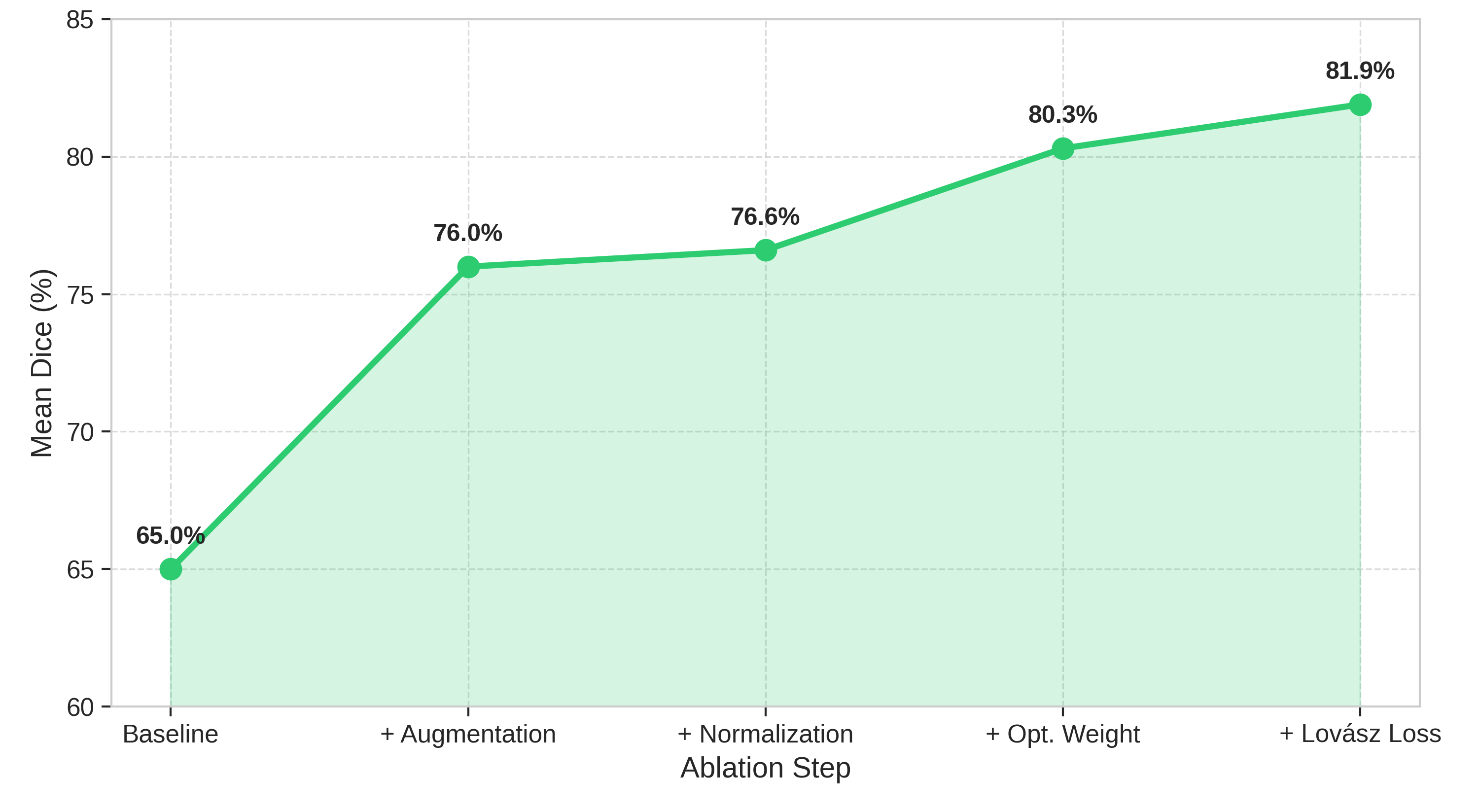
Pure 2D slice-by-slice networks are computationally efficient and datalight, but neglect through plane context risking inconsistent volumetric indices when slice alignment, thickness, or appearance vary between acquisitions [12]. Fully 3D convolutional models address spatial consistency but are memory and computationally heavy, and struggle when data are anisotropic or scarce [13]. Some studies therefore adopt a 2.5D compromise, processing stacks of adjacent slices together to preserve anatomical continuity while maintaining tractable computation [14]. However, such architectures are rarely extended to multi-task reasoning, diagnosis, or report generation. These observations expose key gaps in current practice. First, task design remains fragmented: segmentation, classification, and clinical reporting are handled by separate modules or models. Second, there is little systematic study of how design choices, such as normalization strategy, loss weighting, or architecture depth, influence clinically meaningful outputs like ejection fraction error, misclassification rate, or report fidelity. Third, there is a lack of reproducible end-to-end pipelines: few works trace how improvements in segmentation propagate to diagnosis and clinical indices under varying imaging conditions. In this study, we propose PULSE: a unified transformer-based framework that performs ventricular segmentation, cardiomyopathy classification, and structured clinical report generation directly from short-axis CMR slices. This approach treats cardiac image interpretation as a cohesive, multi-task reasoning problem, from pixels to quantitative indices to narrative summaries.
Deep learning (DL) has emerged as the standard for cardiac image segmentation, with fully convolutional and U-Net derived architectures remaining dominant, and newer attention-or hybrid-based models increasingly explored for richer anatomical context [15,16]. Early U-Net style methods achieved accurate delineation of LV, RV, and myocardium across modalities, as summar
This content is AI-processed based on open access ArXiv data.