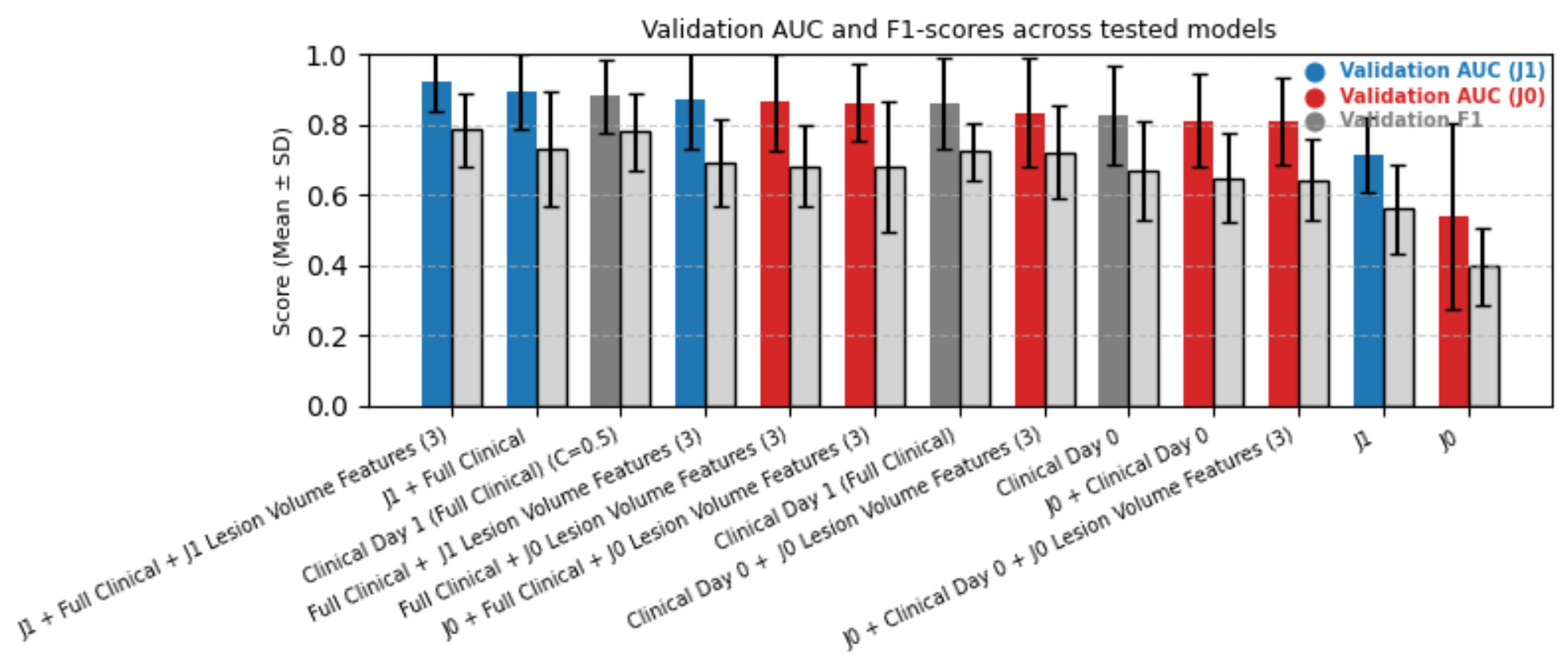
This study compares baseline (J0) and 24-hour (J1) diffusion magnetic resonance imaging (MRI) for predicting three-month functional outcomes after acute ischemic stroke (AIS). Seventy-four AIS patients with paired apparent diffusion coefficient (ADC) scans and clinical data were analyzed. Three-dimensional ResNet-50 embeddings were fused with structured clinical variables, reduced via principal component analysis (<=12 components), and classified using linear support vector machines with eight-fold stratified group cross-validation. J1 multimodal models achieved the highest predictive performance (AUC = 0.923 +/- 0.085), outperforming J0-based configurations (AUC <= 0.86). Incorporating lesion-volume features further improved model stability and interpretability. These findings demonstrate that early post-treatment diffusion MRI provides superior prognostic value to pre-treatment imaging and that combining MRI, clinical, and lesion-volume features produces a robust and interpretable framework for predicting three-month functional outcomes in AIS patients.
Acute ischemic stroke (AIS) is a leading cause of death and long-term disability worldwide [1]. Reliable early prediction of post-stroke functional outcome is critical for treatment planning, rehabilitation, and patient selection in clinical trials [2]. The three-month modified Rankin Scale (mRS) remains the standard measure of disability [3], yet accurate early prediction remains challenging due to heterogeneity in stroke mechanisms, treatment response, and recovery. Such a personalized prognostic model can support treatment decisions, rehabilitation planning, and adaptive trial enrollment, addressing a major unmet clinical need in stroke management.
Diffusion-weighted MRI (DWI) and its quantitative map, the apparent diffusion coefficient (ADC), are sensitive to early ischemic injury [4]. Baseline (J0) ADC reflects the infarct core, while day-1 (J1) imaging captures infarct evolution, reperfusion, and secondary injury processes [5,6]. Although both time points are acquired routinely, their relative prognostic value for long-term functional outcome remains unclear.
Traditional models combining clinical scores such as age, NIHSS (National Institutes of Health Stroke Score), and prestroke mRS achieve moderate predictive accuracy (AUC ≈ 0.75-0.80) [7], but underuse imaging biomarkers reflecting dynamic tissue changes. Deep learning enables automated extraction of high-dimensional image representations [8,9], and recent multimodal models integrating imaging and clinical data have improved prediction [10,11]. However, most prior studies focus solely on baseline imaging.
To our knowledge, this is the first study to directly compare baseline (J0) and early follow-up (J1) diffusion MRI for predicting three-month functional outcome in AIS using a unified multimodal deep-embedding framework. We hypothesize that post-treatment (J1) imaging, combined with clinical and lesion-volume features, can improve predictive performance and interpretability. Our framework integrates 3D ResNet-derived ADC embeddings with structured clinical variables and applies Principal Component Analysis (PCA) for dimensionality reduction and linear SVM classification to enable transparent, data-efficient outcome prediction.
Functional outcome prediction after AIS has relied on clinical scoring and regression models. Baseline neurological severity (NIHSS), age, and pre-stroke mRS remain the strongest predictors of three-month outcomes [2]. Logistic or Cox models using these variables achieve moderate accuracy (AUC ≈ 0.75-0.80) [7,12], but lack sensitivity to imaging biomarkers reflecting infarct evolution or treatment response.
Machine learning and deep learning methods can further improve prediction by combining imaging and clinical features [13,10,8]. CNNs and 3D architectures enable automated feature extraction from ADC maps [14,15], and multimodal fusion models show superior accuracy [11]. Yet most arXiv:2512.02088v1 [eess.IV] 1 Dec 2025 To mitigate redundancy in deep embeddings, PCA often offers efficient dimensionality reduction and improved generalization [16]. Our study builds on this by comparing J0 and J1 imaging, integrating deep volumetric and clinical features, and employing PCA-based fusion with linear SVM classification for interpretable AIS outcome prediction.
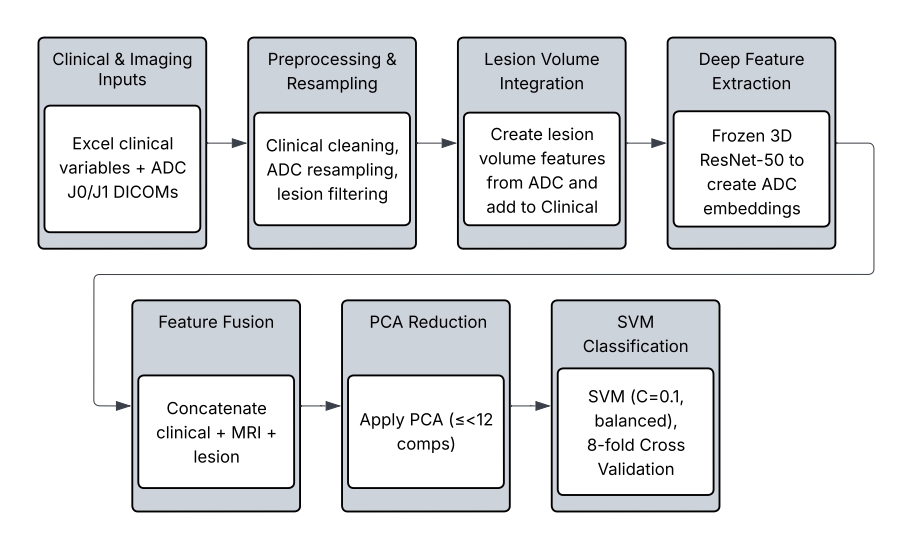
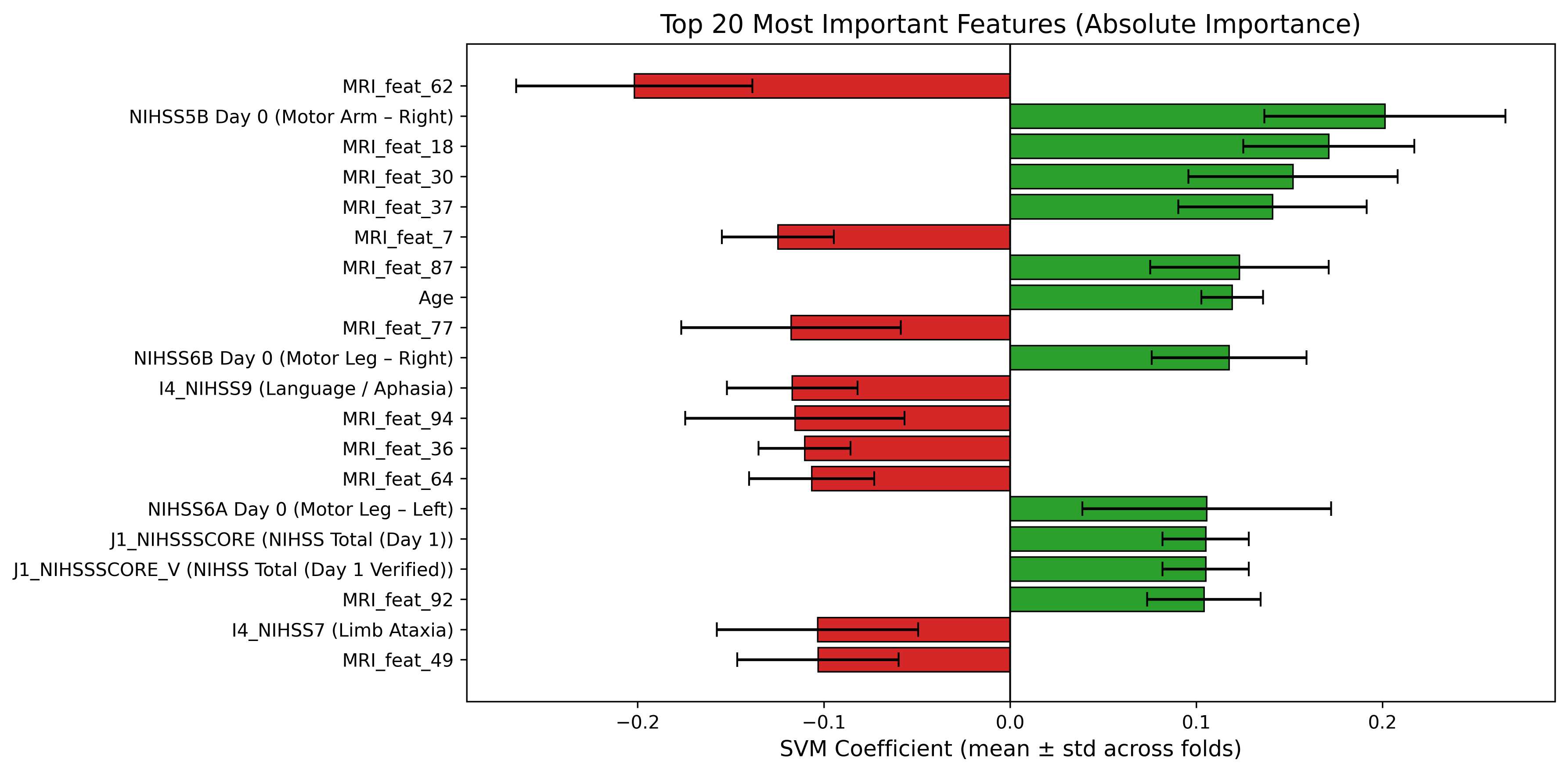
74 patients with AIS who underwent DWI MRI at J0 and J1 were retrospectively selected from a prospective registry under institutional approval. Inclusion required paired ADC volumes, complete clinical records (NIHSS subscores, prestroke mRS), and available three-month mRS. Demographics, vascular risk factors, pre-mRS, and NIHSS (J0, J1) were included. Both anterior and posterior strokes were included; outcomes were 55.4% favorable (mRS ≤ 1) and 44.6% unfavorable (mRS > 1). Missing values were imputed using median substitution, and all data were anonymized. The overall data processing and modeling workflow is illustrated in Figure 2, which summarizes each step from image preprocessing and lesion extraction to feature fusion and classification.
ADC maps reconstructed from 1.5 T and 3 T DWI were resampled via trilinear interpolation to a unified 3D resolution of 24 × 256 × 256 voxels.
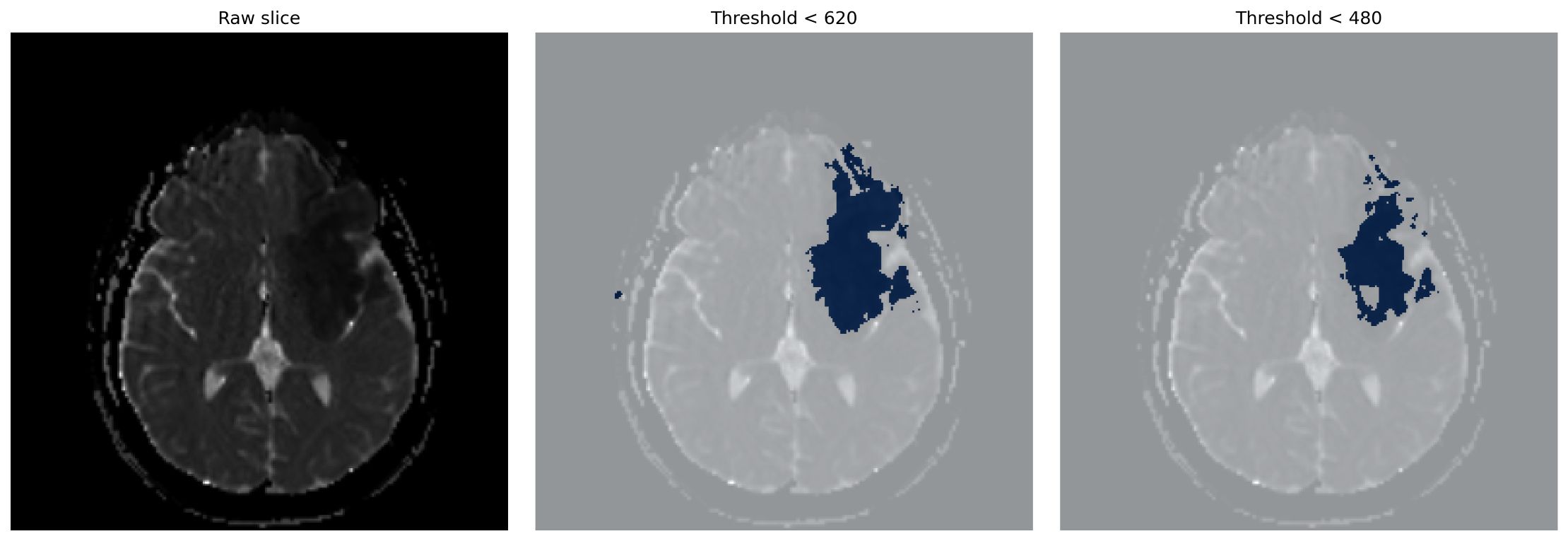
Lesion-like regions were segmented using percentilebased intensity thresholds (480 and 620 ×10 -6 mm 2 /s), followed by morphological filtering and removal of small components (<150 voxels). An example of the thresholding process at different intensity levels is shown in Figure 1, where (left) shows the raw slice, (middle) shows the result for threshold < 620, and (right) shows the result for threshold < 480. Lesion volume (V lesion = N voxels × V voxel ) was log-transformed and concatenated with clinical data.
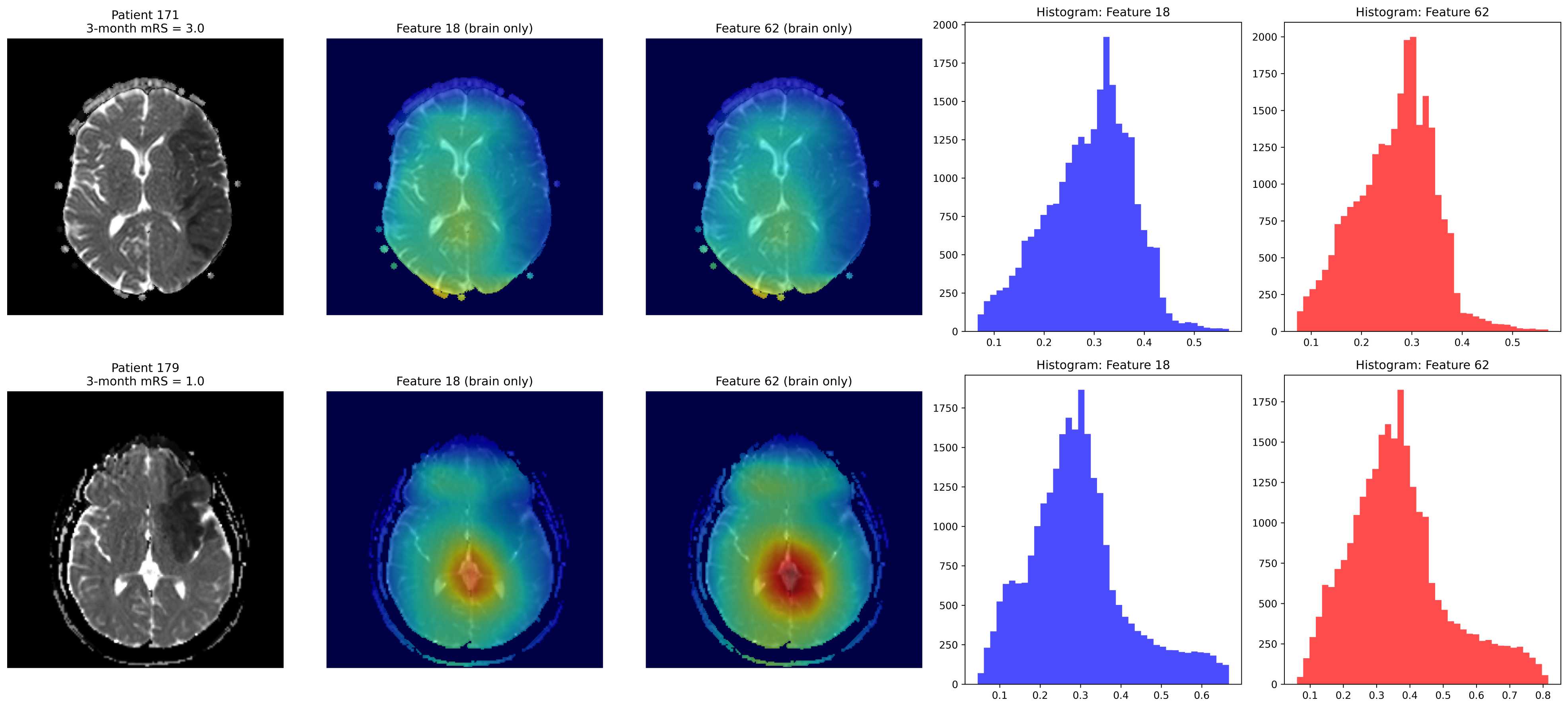
Volume features are extracted with a pretrained 3D ResNet-50 (MedicalNet [17]) implemented in MONAI [18] (Medical Open Network for AI). Separate branches processed J0 and J1 ADC input, with global average pooling producing 2048-D embeddings projected to 32-256 units. Networks ope
This content is AI-processed based on open access ArXiv data.