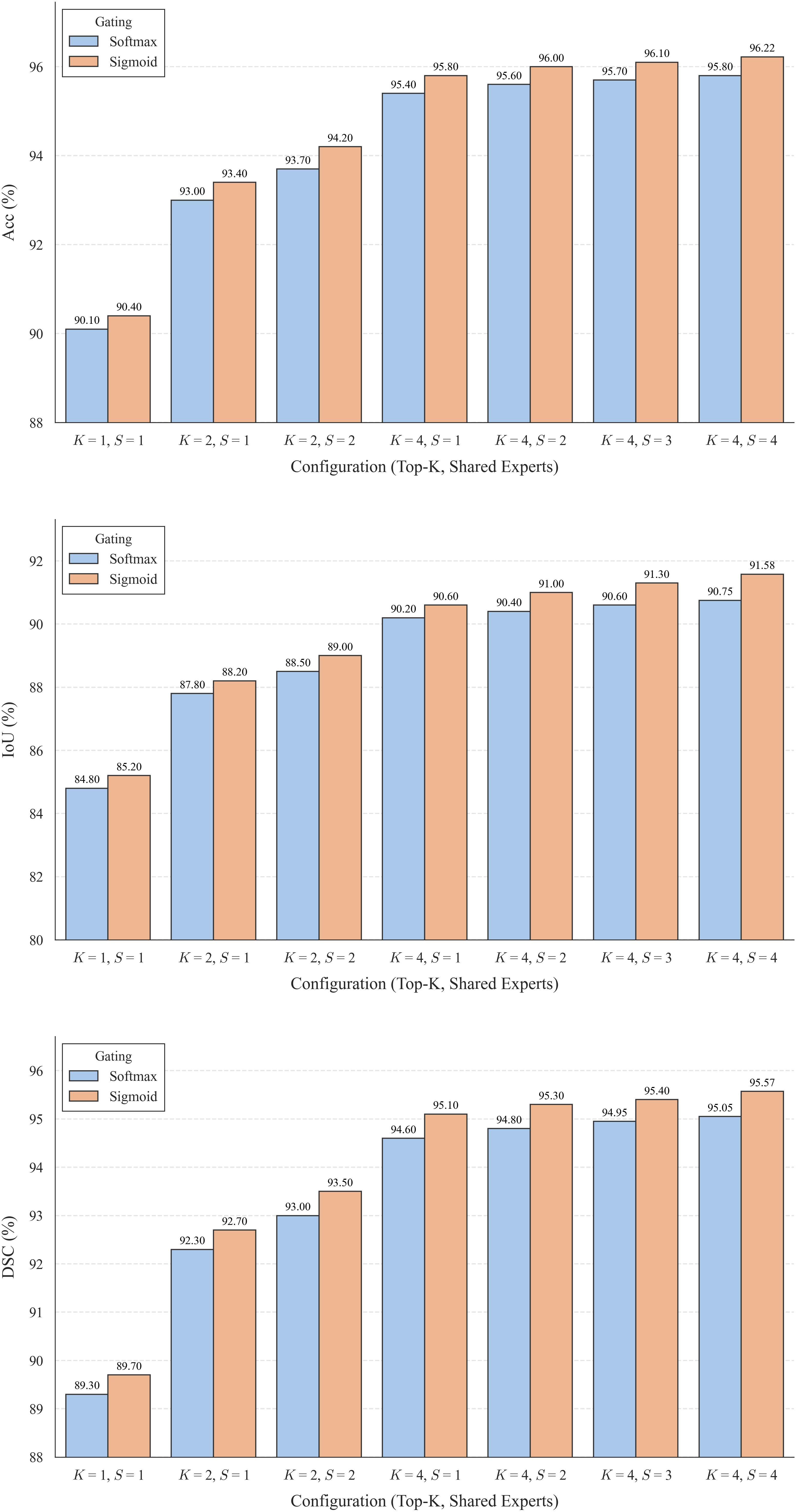
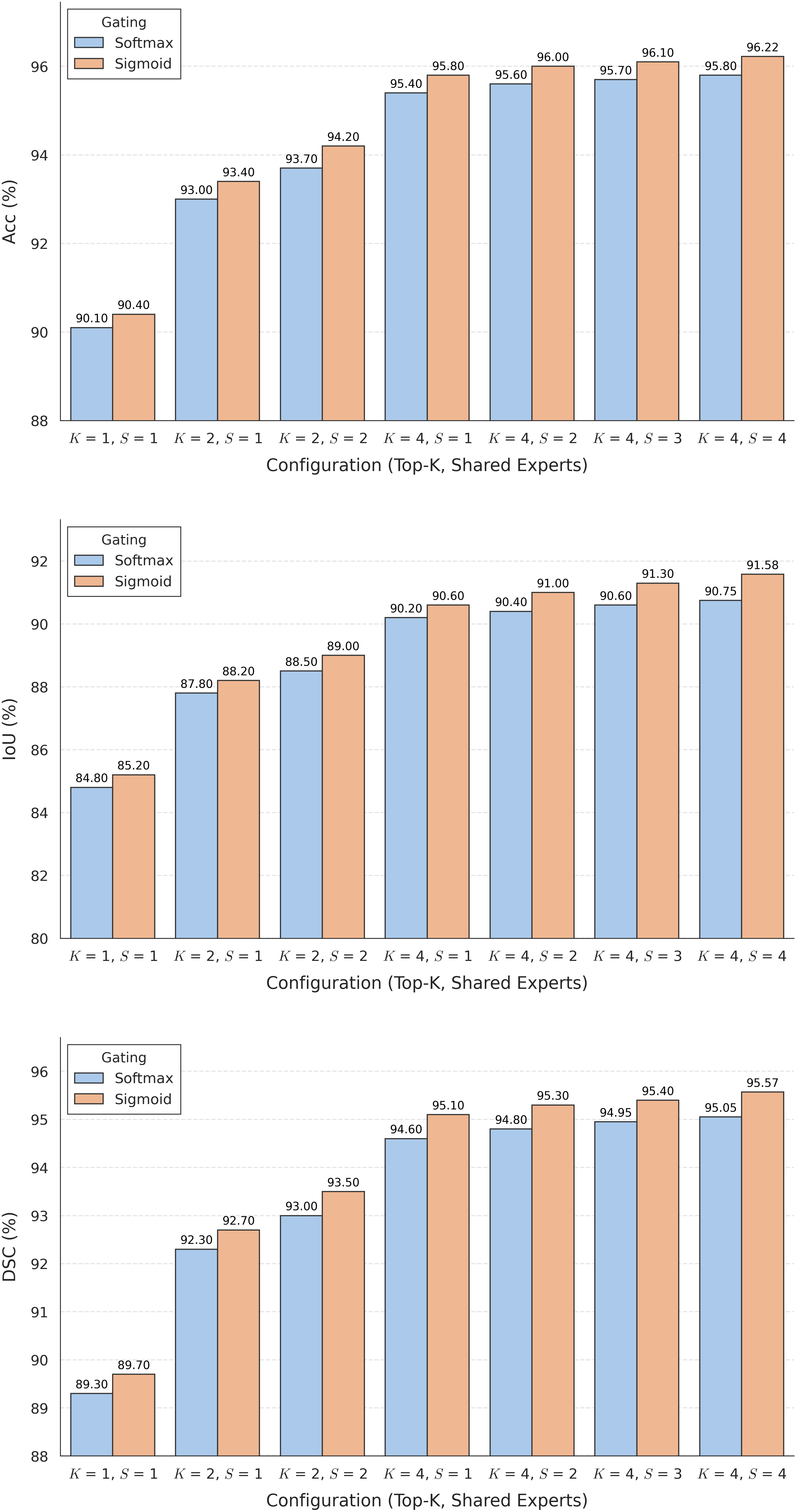
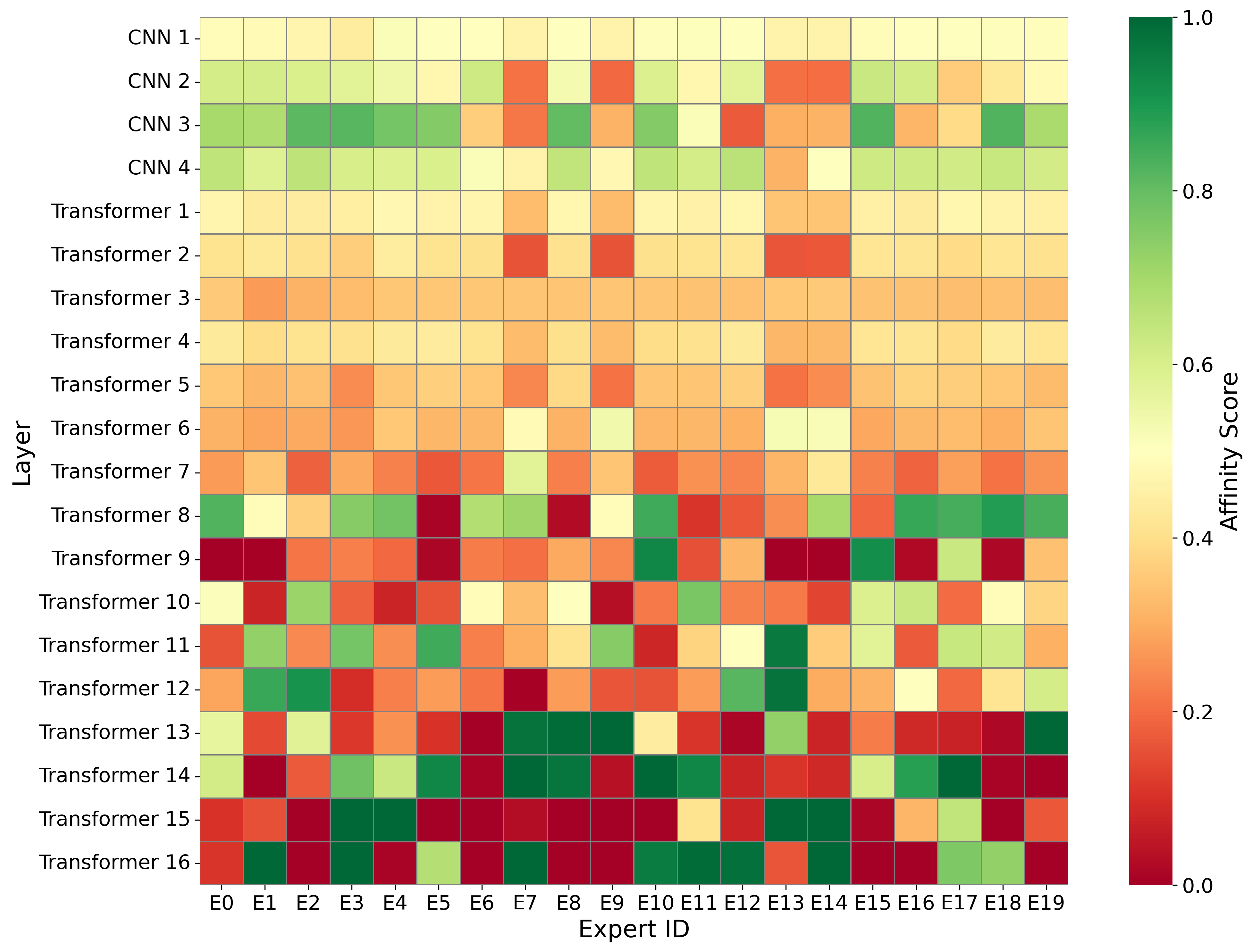
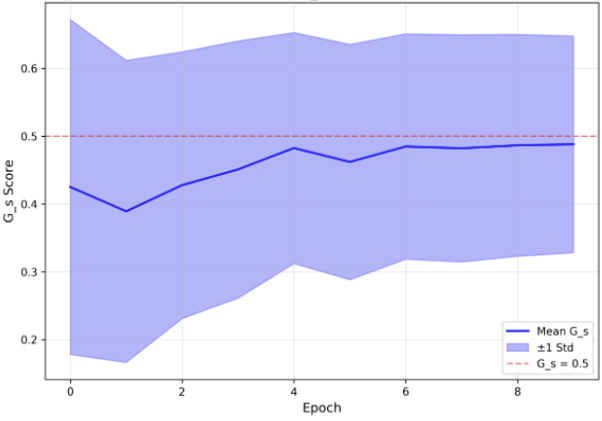
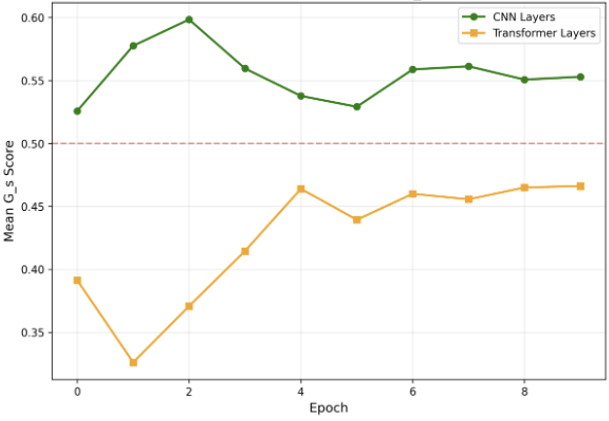
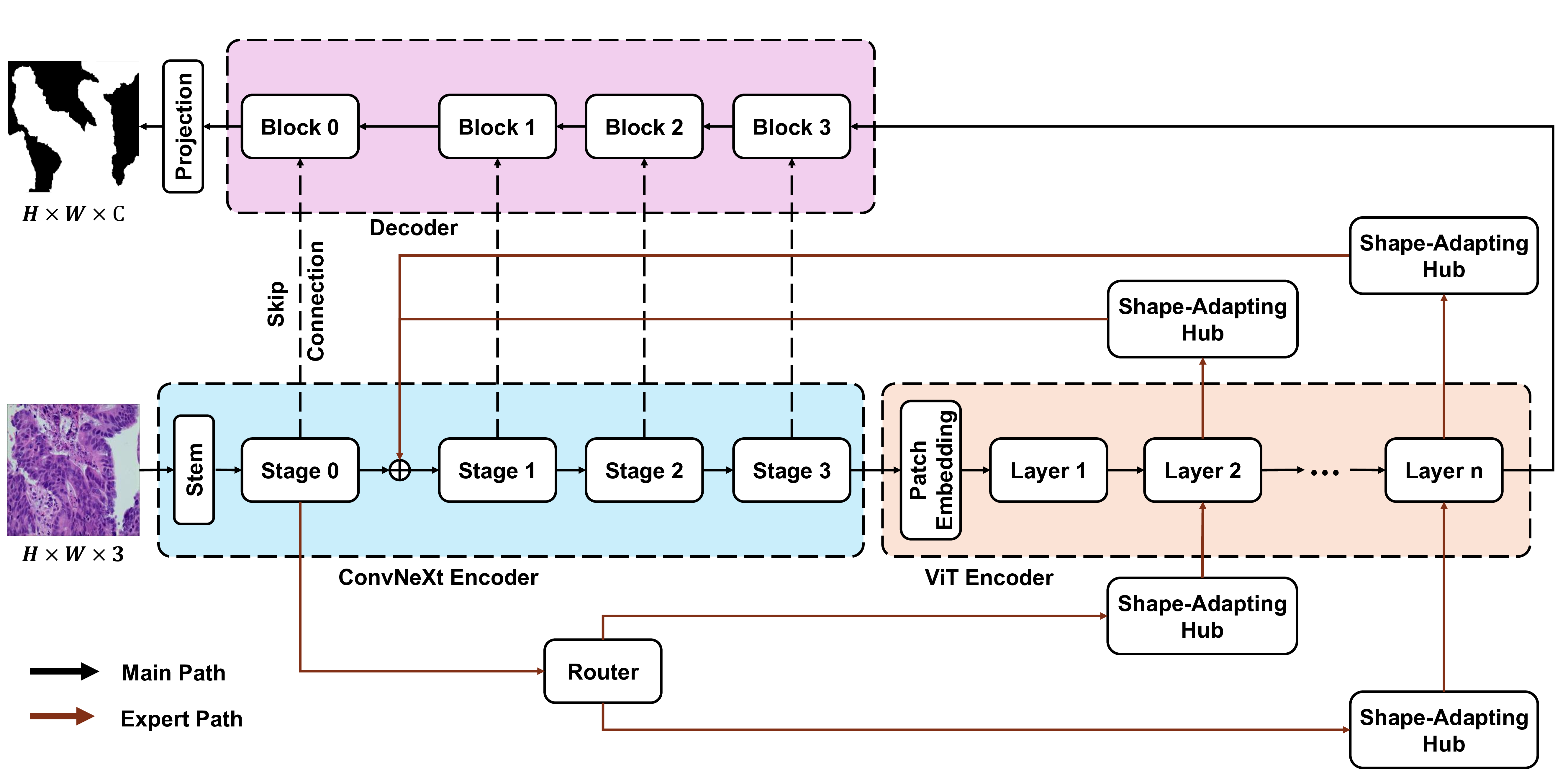
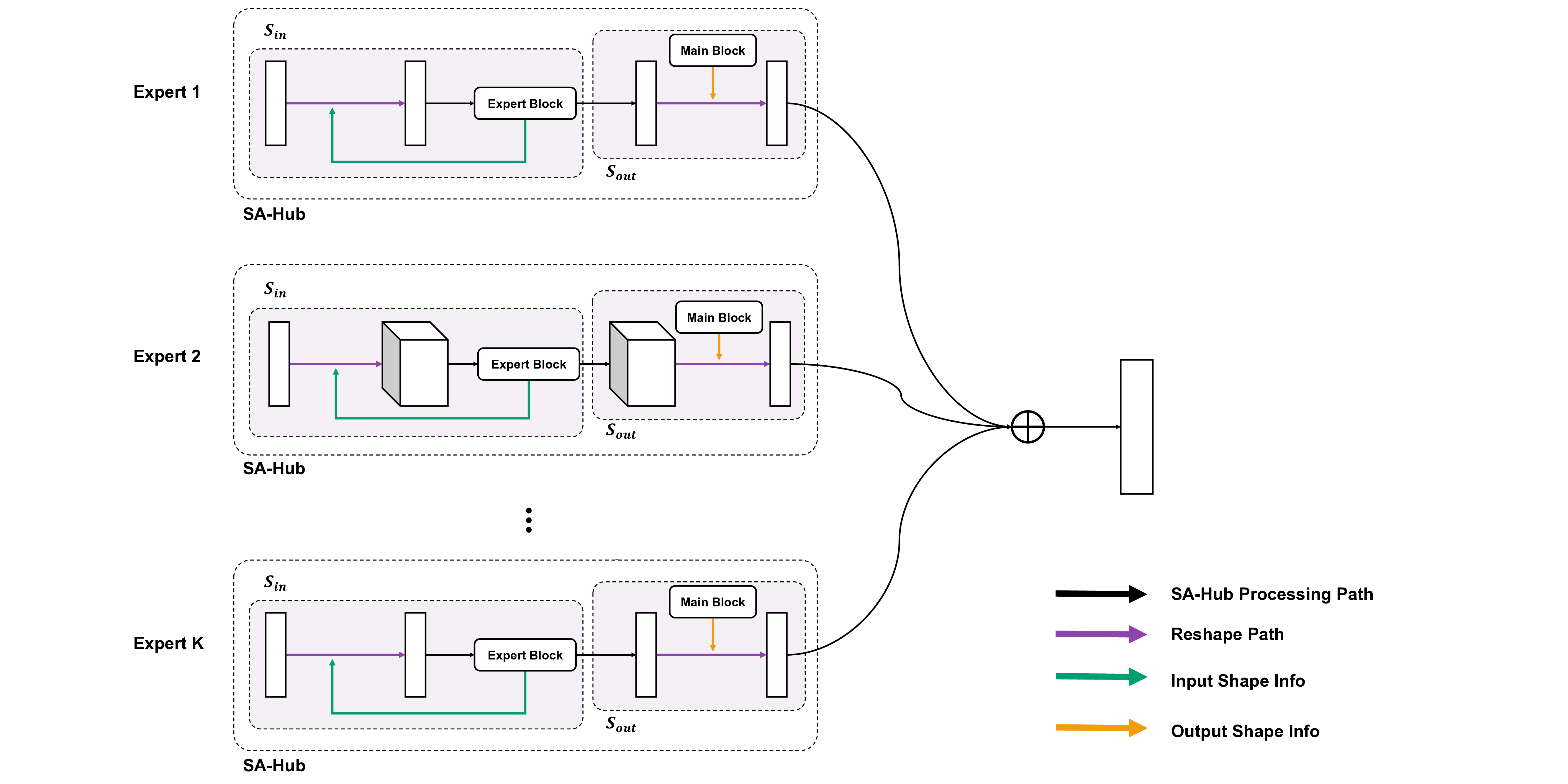
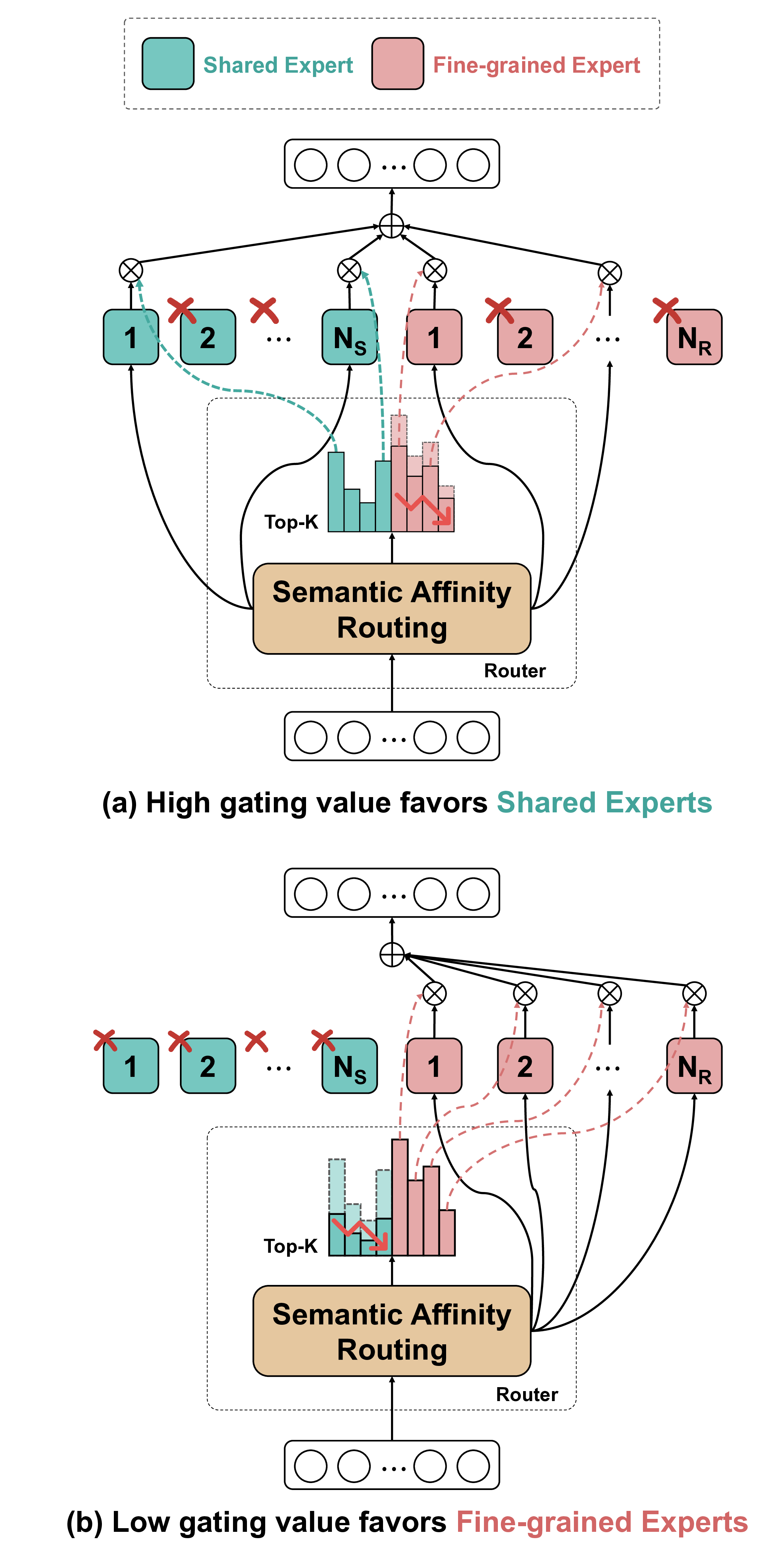
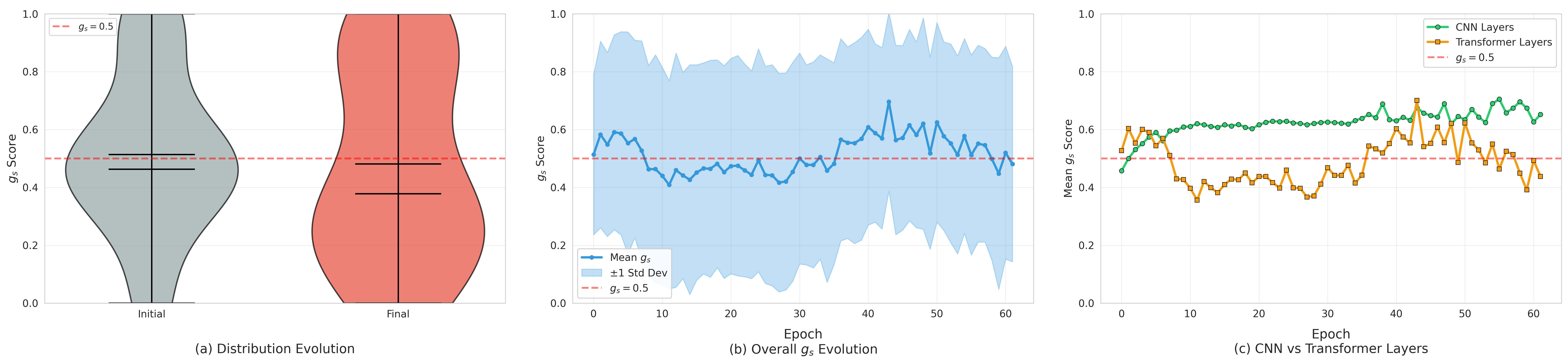
The substantial diversity in cell scale and form remains a primary challenge in computer-aided cancer detection on gigapixel Whole Slide Images (WSIs), attributable to cellular heterogeneity. Existing CNN-Transformer hybrids rely on static computation graphs with fixed routing, which consequently causes redundant computation and limits their adaptability to input variability. We propose Shape-Adapting Gated Experts (SAGE), an input-adaptive framework that enables dynamic expert routing in heterogeneous visual networks. SAGE reconfigures static backbones into dynamically routed expert architectures. SAGE's dual-path design features a backbone stream that preserves representation and selectively activates an expert path through hierarchical gating. This gating mechanism operates at multiple hierarchical levels, performing a two-level, hierarchical selection between shared and specialized experts to modulate model logits for Top-K activation. Our Shape-Adapting Hub (SA-Hub) harmonizes structural and semantic representations across the CNN and the Transformer module, effectively bridging diverse modules. Embodied as SAGE-UNet, our model achieves superior segmentation on three medical benchmarks: EBHI, DigestPath, and GlaS, yielding state-of-the-art Dice Scores of 95.57%, 95.16%, and 94.17%, respectively, and robustly generalizes across domains by adaptively balancing local refinement and global context. SAGE provides a scalable foundation for dynamic expert routing, enabling flexible visual reasoning.
The foundation of digital pathology lies in computer-aided detection of malignant tissue within gigapixel WSIs, enabling rapid and quantitative diagnosis. In colorectal cancer, accurately characterizing the tumor's morphology is crucial for prompt diagnosis, classification, and treatment planning. However, translating such visually complex and heterogeneous tissue structures into computational under-standing remains highly challenging. While Convolutional Neural Networks (CNNs) [1] excel at capturing fine-grained local features, such as cell boundaries and textures, Vision Transformers (ViTs) [15] have emerged as a powerful paradigm for modeling long-range spatial dependencies and global context. Nevertheless, substantial variability in tissue appearance, ranging from homogeneous normal tissues to complex and subtly textured malignant patterns, combined with the large resolution of WSIs, pushes current models beyond their representational and computational limits. Existing models, including U-Net variants and hybrid CNN-Transformer architectures, utilize a static computational graph. This forces uniform processing for all input segments, an inefficient and suboptimal approach that over-processes simple regions while under-modeling complex ones. Furthermore, the fixed interaction between CNN and Transformer blocks prevents the adaptive leveraging of each paradigm's strengths based on input characteristics.
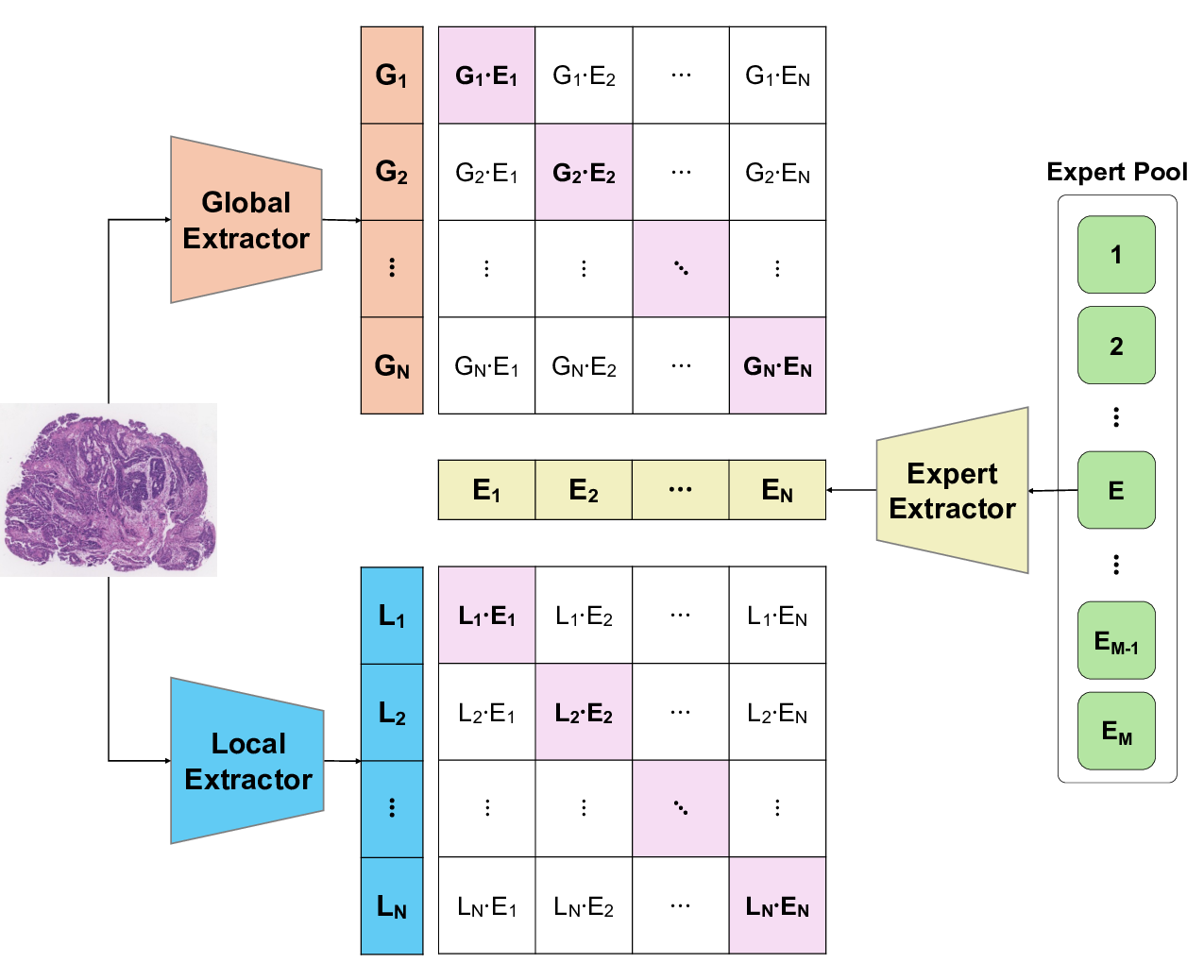
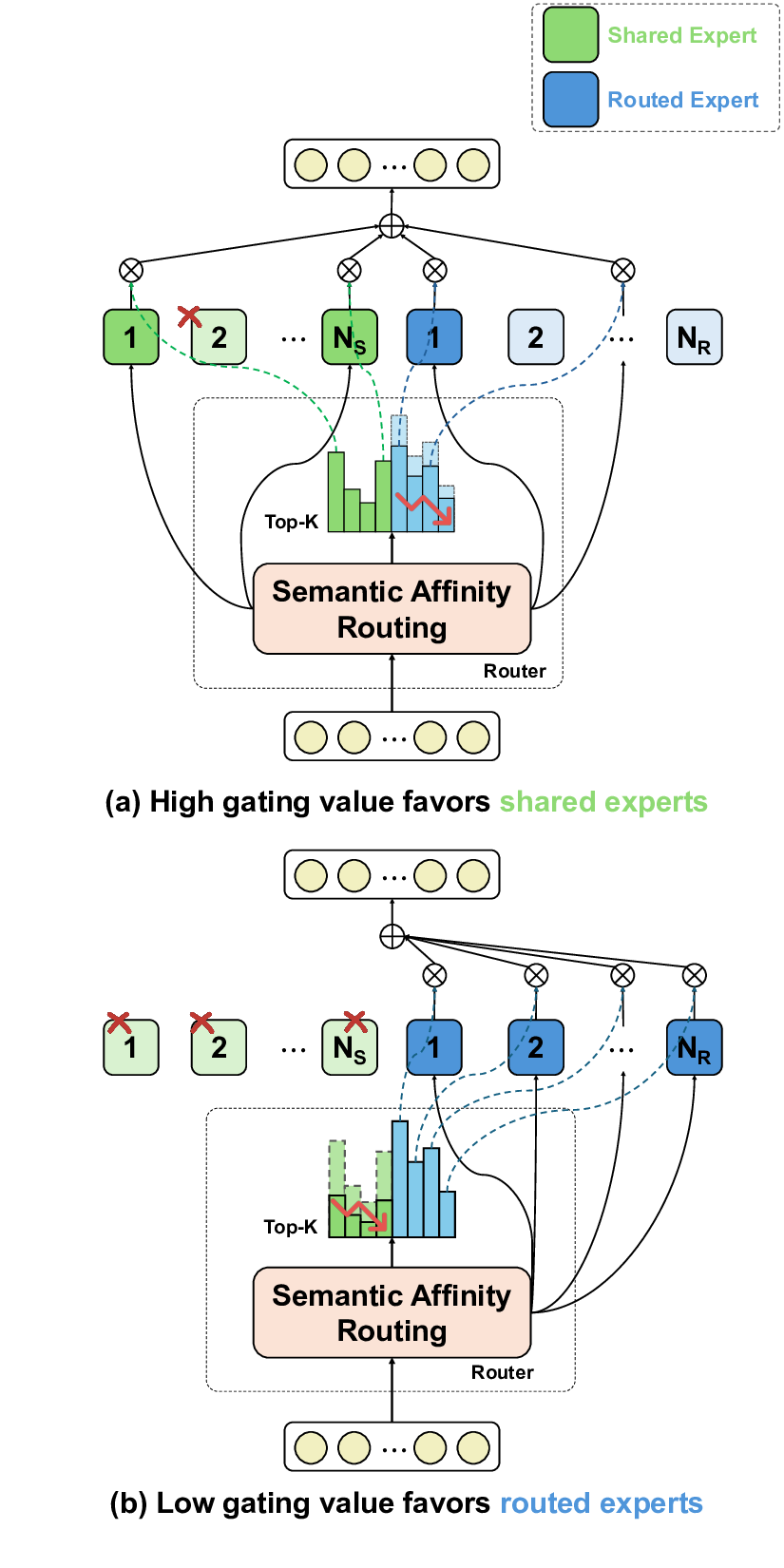
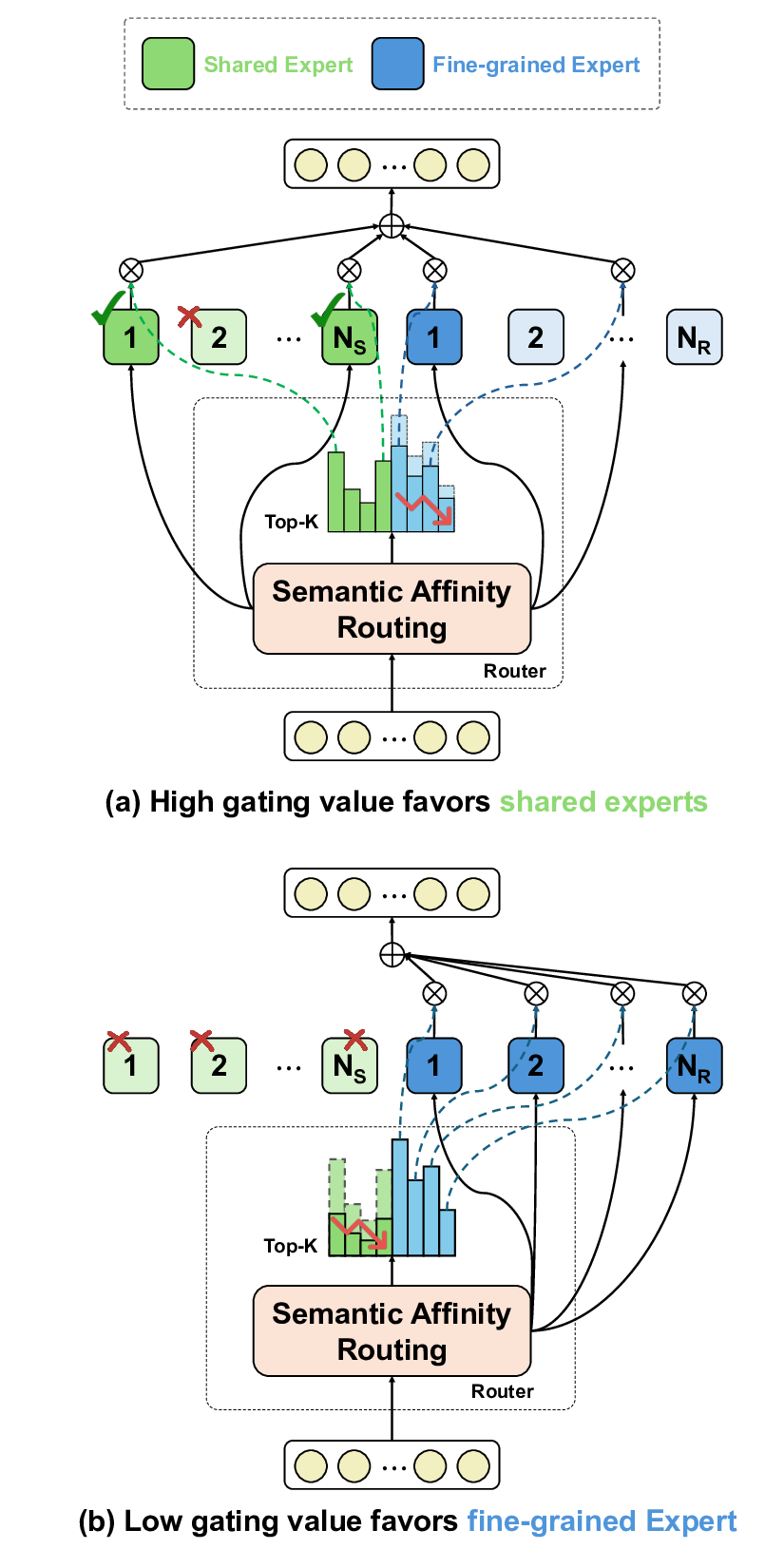
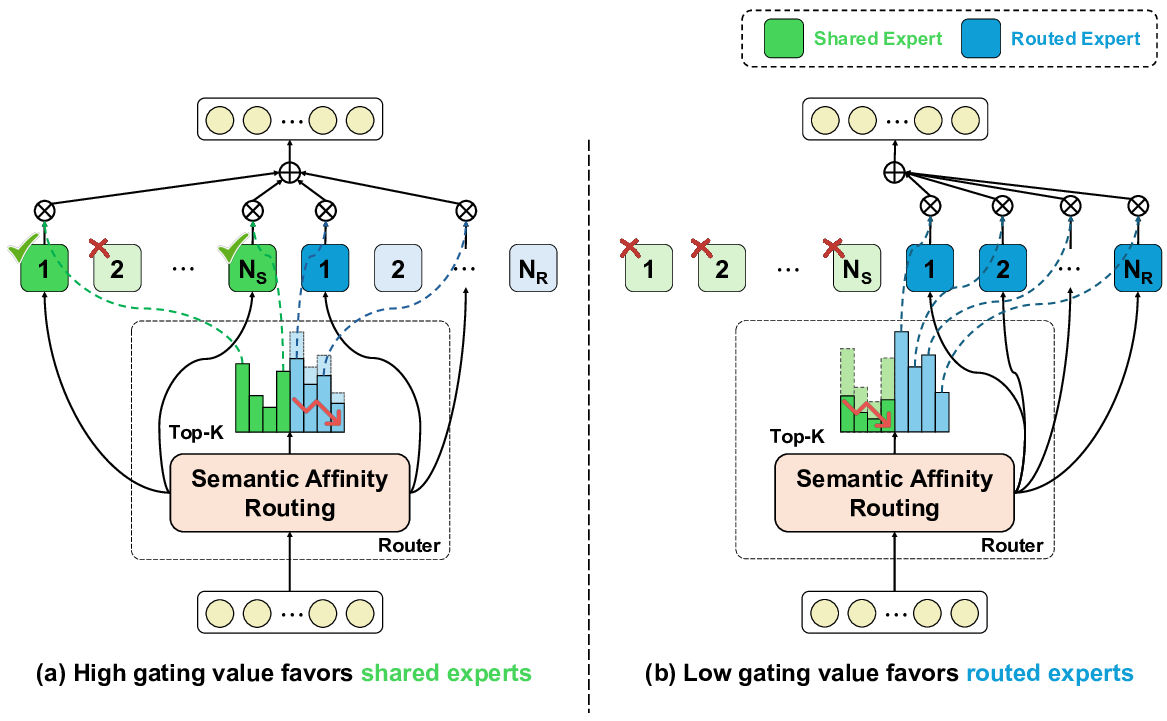
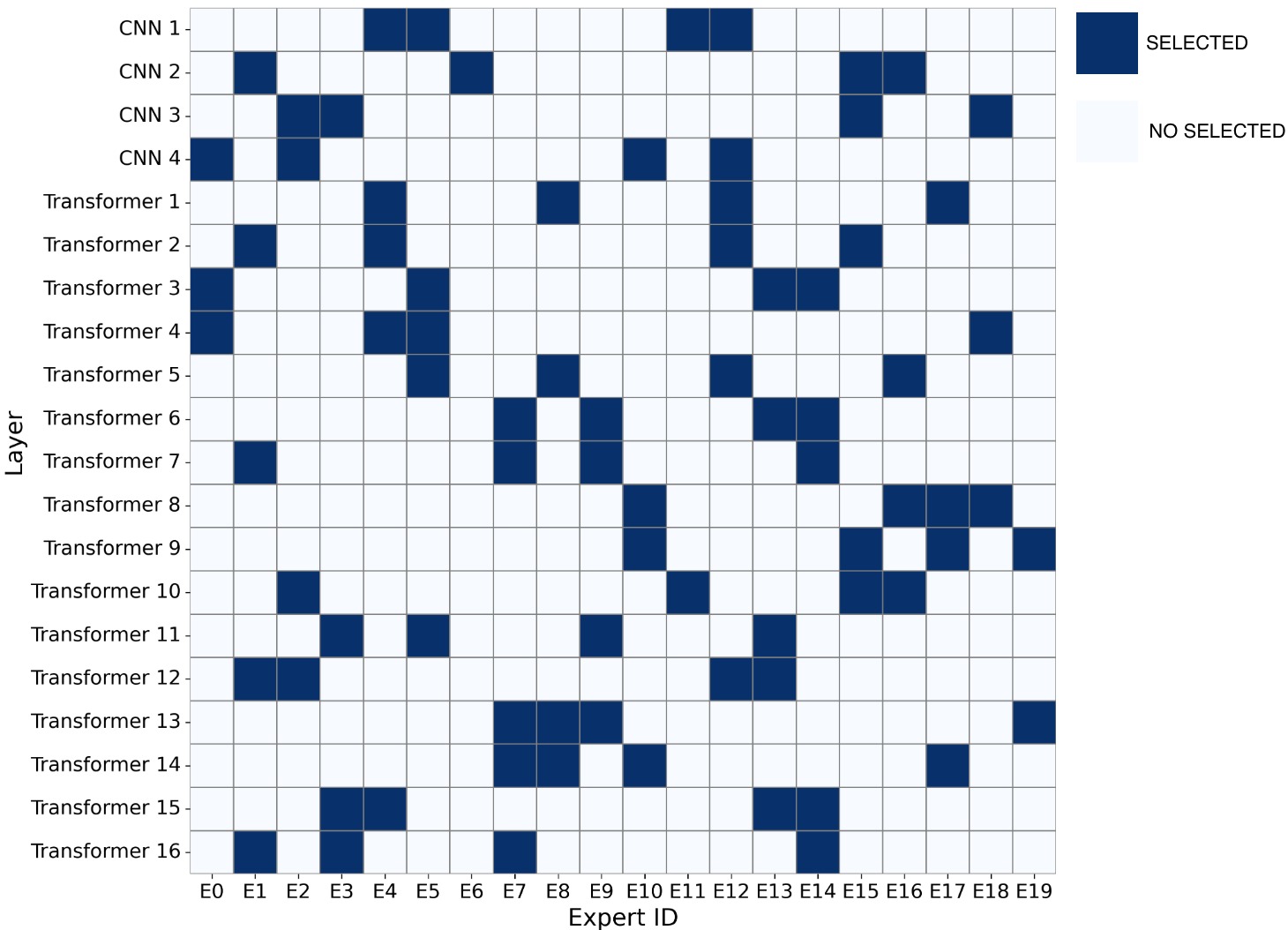
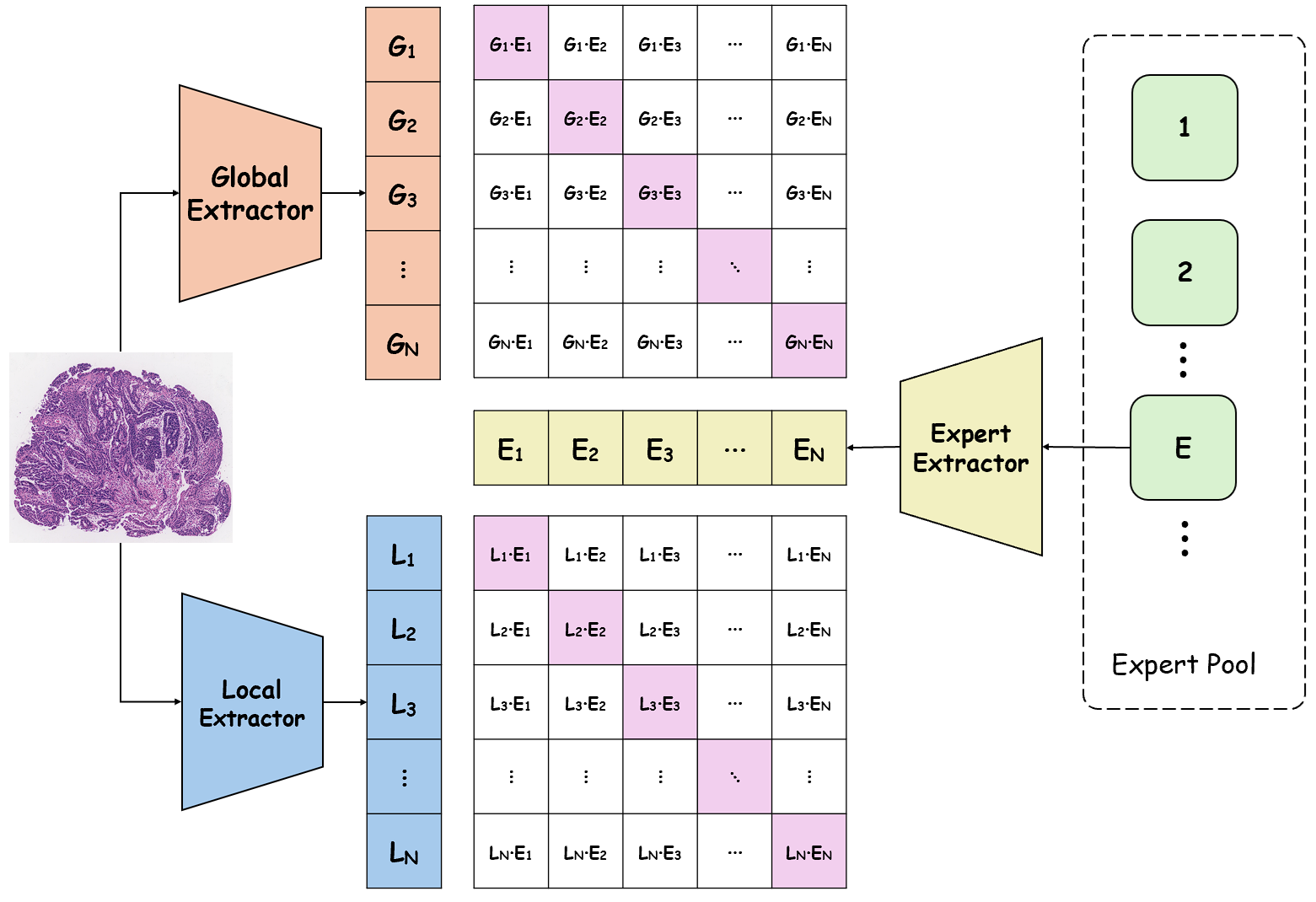
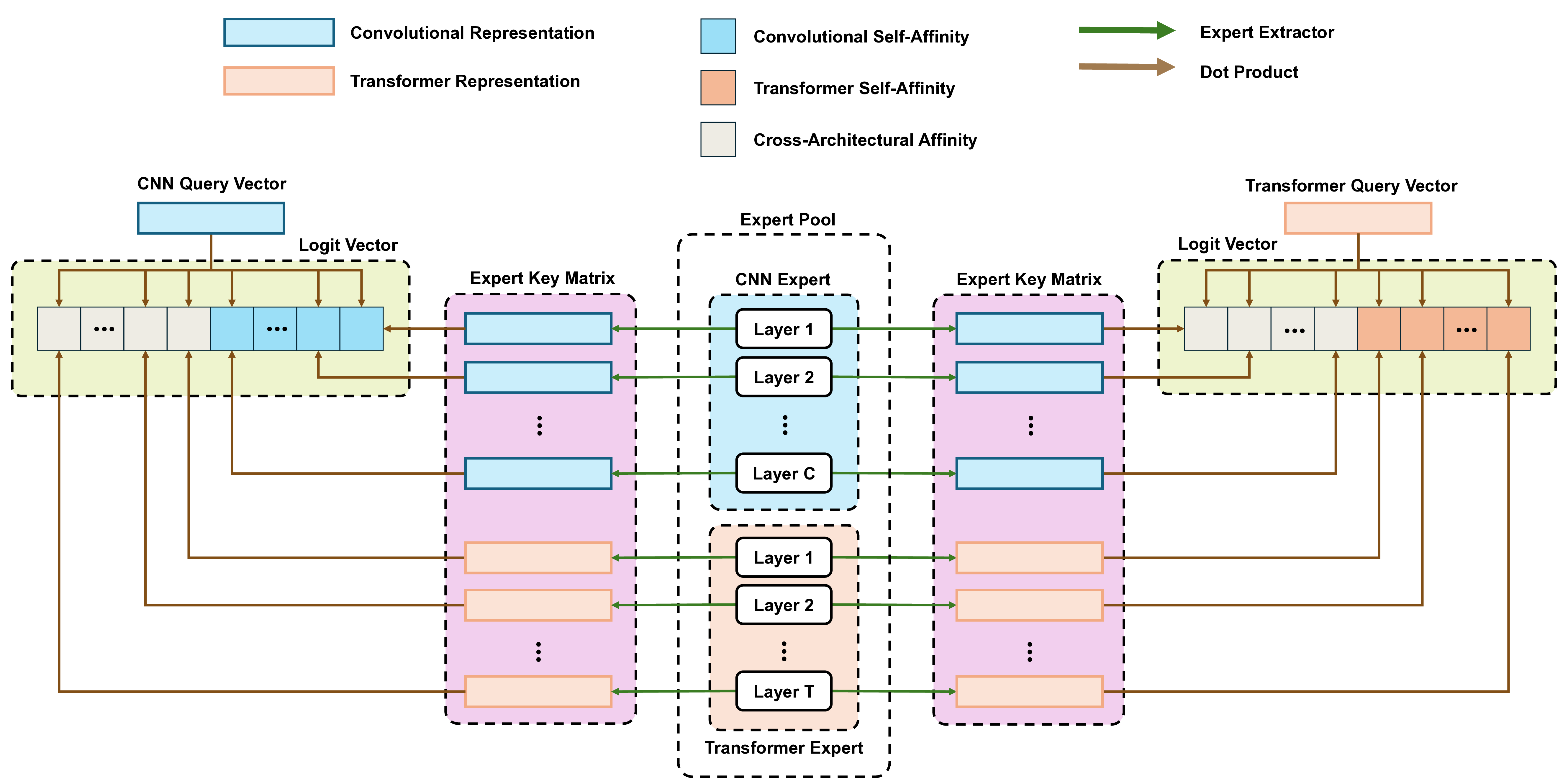
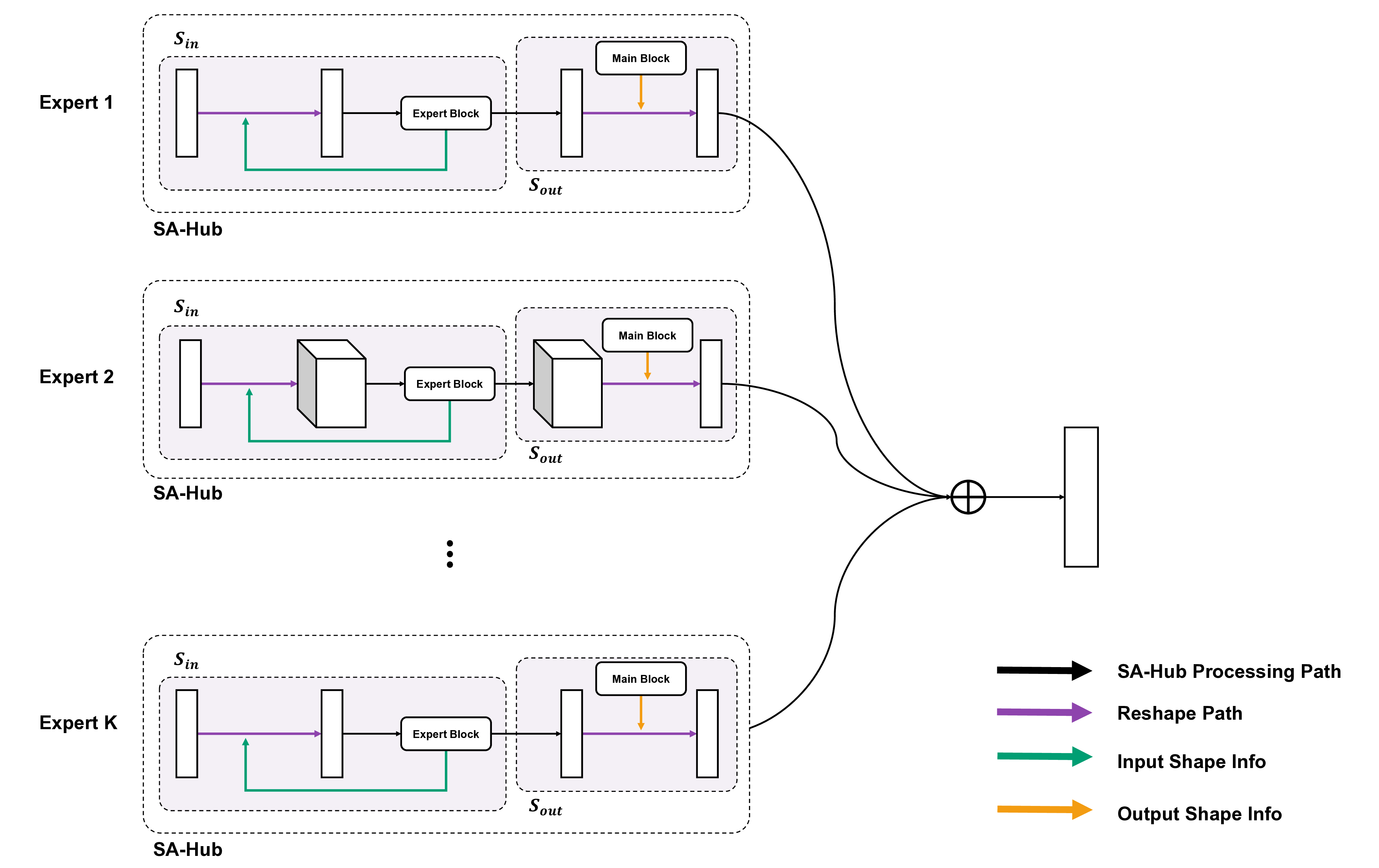
To address these limitations, we propose Shape-Adapting Gated Experts (SAGE), a dynamic, input-adaptive framework that converts any static backbone into a flexible dual-path system. SAGE introduces a dual-path layer architecture, comprising a main path that preserves the original backbone for stability and the reuse of pre-trained knowledge, and an expert path that selectively activates specialized computations conditioned on the input. The model learns to combine these two paths, dynamically determining the degree of expert-driven refinement required at each stage. In contrast to traditional flat Mixture-of-Experts (MoE) routing, SAGE utilizes a hierarchical gating mechanism that initially differentiates between shared and finegrained experts, subsequently executing top-K routing to allocate computational resources efficiently. At the top level, a gate selects between general-purpose shared experts and special-purpose fine-grained experts, which are the underlying blocks of the backbone. The final top-K selection is then directed toward the optimal computational paradigm. To facilitate communication within this heterogeneous expert pool, we introduce the Shape-Adapting Hub (SA-Hub). This module dynamically adapts feature tensors to bridge the architectural gap between different types of experts, such as CNNs and Transformers.
To summarize, this work makes contributions as follows: • We propose a dual-path architecture that enables static backbones to become dynamic, allowing for inputdependent computation and the adaptive integration of local and global context by routing features among diverse experts, including the original backbone blocks. • We design a hierarchical expert routing system that distinguishes between shared and fine-grained experts, ensuring more adaptive and context-aware selection. • We introduce a lightweight module that enables CNNs, Transformers, and other models to align their feature rep-resentations dynamically.
Medical Image Segmentation. Image segmentation in the medical domain has been at the forefront of computational pathology, allowing detailed analysis of cellular and tissue-level morphology. Traditional approaches conducted based on pixel intensities, region growing, and/or evolution of boundaries are inherently very sensitive to noise and morphology variation. The emergence of deep learning has significantly influenced image segmentation, and its application was first proposed with U-Net [27], with subsequent improvements over enhanced backbones such as ResNet [12], EfficientNet [32], ConvNeXt [19], and others with hierarchical designs such as U-Net++ [40]. Recent research further emphasizes data efficiency and crossdomain generalization. Foundation models such as Med-SAM [21] and SAM-Med2D/3D [31] leverage large-scale pretraining with task-guided segmentation to yield transferable representations across modalities. However, domain heterogeneity and scarce annotations hinder robust adaptation to histopathology. Semi-supervised approaches, including C2GMatch [25], mitigate this issue via weak-tostrong consistency learning, reflecting a shift toward dataefficient, domain-adaptive segmentation. Hybrid U-Net Architectures. Recent advances in medical image segmentation, however, have seen growth in efforts to enlarge the receptive fields and allow global context comprehension, keeping computational overhead in check. The models based on the transformer structure, such as TransUNet [5], Swin-UNet [2], and SegFormer [37], can understand long State-space models, such as U-Mamb
This content is AI-processed based on open access ArXiv data.