Improving Gene Trees without more data
📝 Original Info
- Title: Improving Gene Trees without more data
- ArXiv ID: 2511.03692
- Date: 2025-11-05
- Authors: 정보 없음 (논문에 명시된 저자 정보가 제공되지 않음)
📝 Abstract
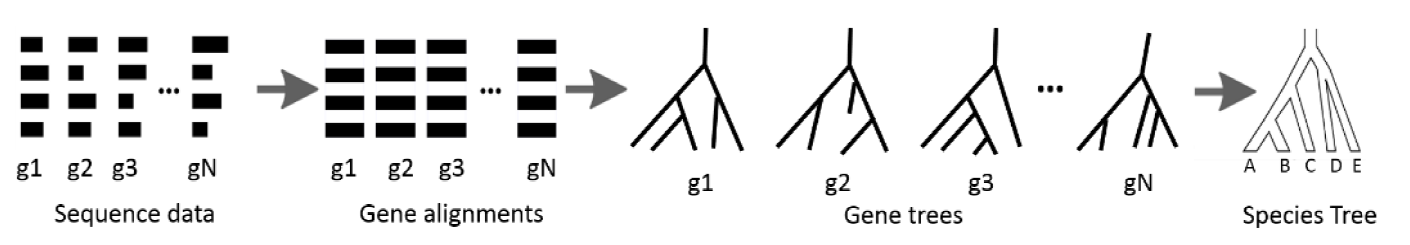
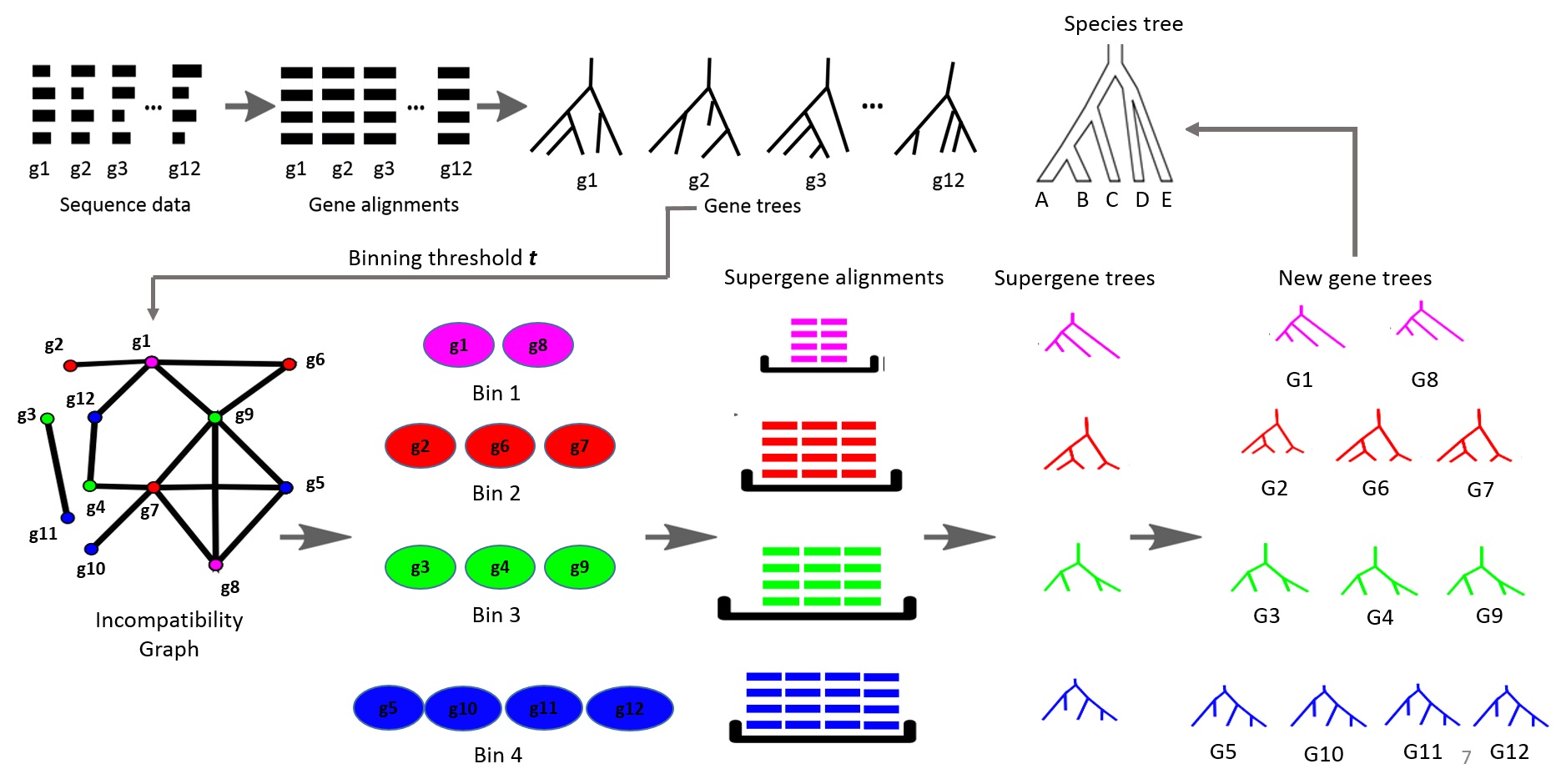
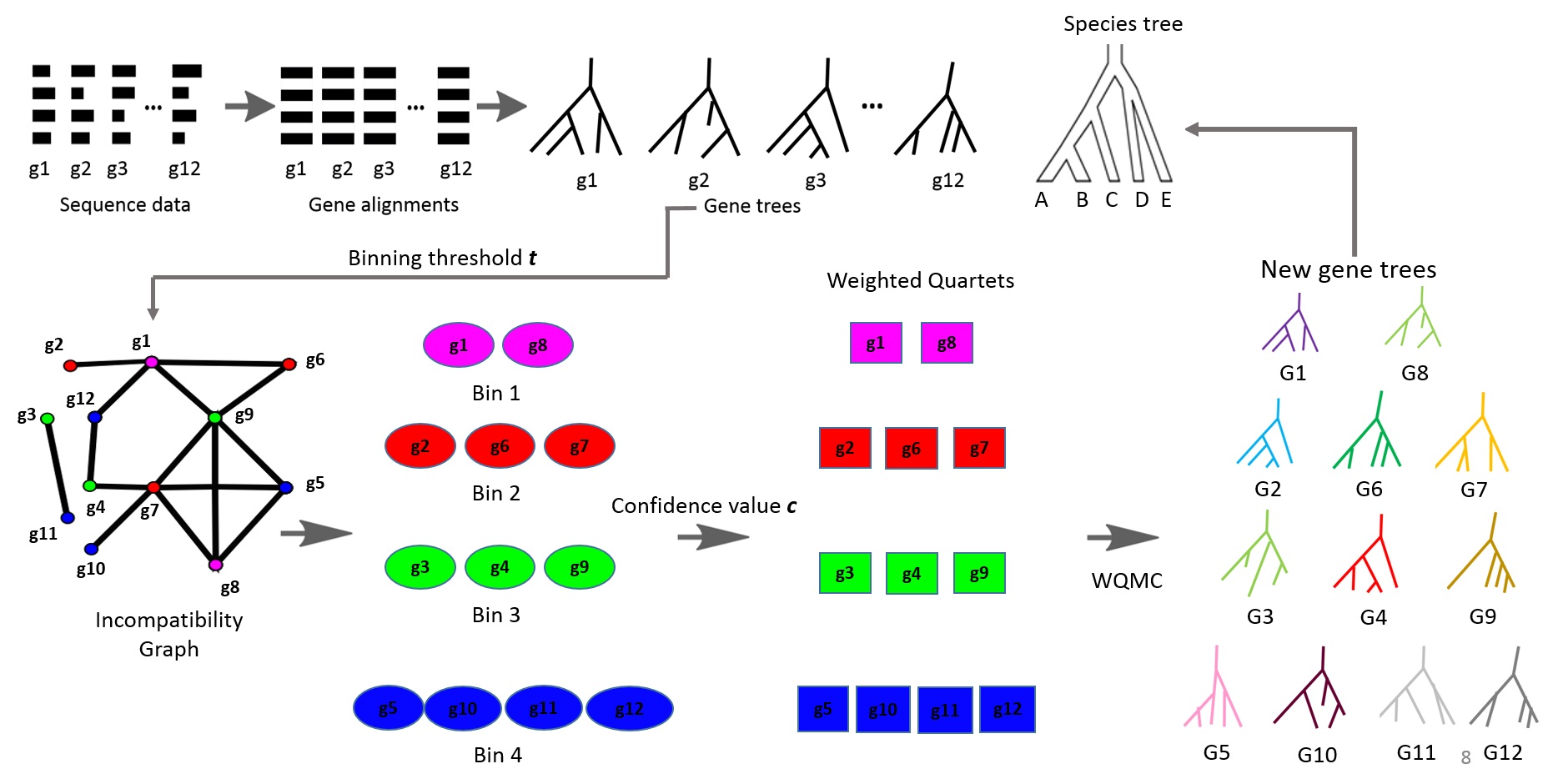
Estimating species and gene trees from sequence data is challenging. Gene tree estimation is often hampered by low phylogenetic signal in alignments, leading to inaccurate trees. Species tree estimation is complicated by incomplete lineage sorting (ILS), where gene histories differ from the species' history. Summary methods like MP-EST, ASTRAL2, and ASTRID infer species trees from gene trees but suffer when gene tree accuracy is low. To address this, the Statistical Binning (SB) and Weighted Statistical Binning (WSB) pipelines were developed to improve gene tree estimation. However, previous studies only tested these pipelines using multi-locus bootstrapping (MLBS), not the BestML approach. This thesis proposes a novel pipeline, WSB+WQMC, which shares design features with the existing WSB+CAML pipeline but has other desirable properties and is statistically consistent under the GTR+MSC model. This study evaluated WSB+WQMC against WSB+CAML using BestML analysis on various simulated datasets. The results confirmed many trends seen in prior MLBS analyses. WSB+WQMC substantially improved gene tree and species tree accuracy (using ASTRAL2 and ASTRID) on most datasets with low, medium, and moderately high ILS levels. In a direct comparison, WSB+WQMC computed less accurate trees than WSB+CAML under certain low and medium ILS conditions. However, WSB+WQMC performed better or at least as accurately as WSB+CAML on all datasets with moderately high and high ILS. It also proved better for estimating gene trees on some medium and low ILS datasets. Thus, WSB+WQMC is a promising alternative to WSB+CAML for phylogenetic estimation, especially in the presence of low phylogenetic signal.💡 Deep Analysis
📄 Full Content
📸 Image Gallery

Reference
This content is AI-processed based on open access ArXiv data.