Diffusion Models at the Drug Discovery Frontier: A Review on Generating Small Molecules versus Therapeutic Peptides
📝 Original Info
- Title: Diffusion Models at the Drug Discovery Frontier: A Review on Generating Small Molecules versus Therapeutic Peptides
- ArXiv ID: 2511.00209
- Date: 2025-10-31
- Authors: 제공된 정보에 저자 명단이 포함되어 있지 않습니다.
📝 Abstract
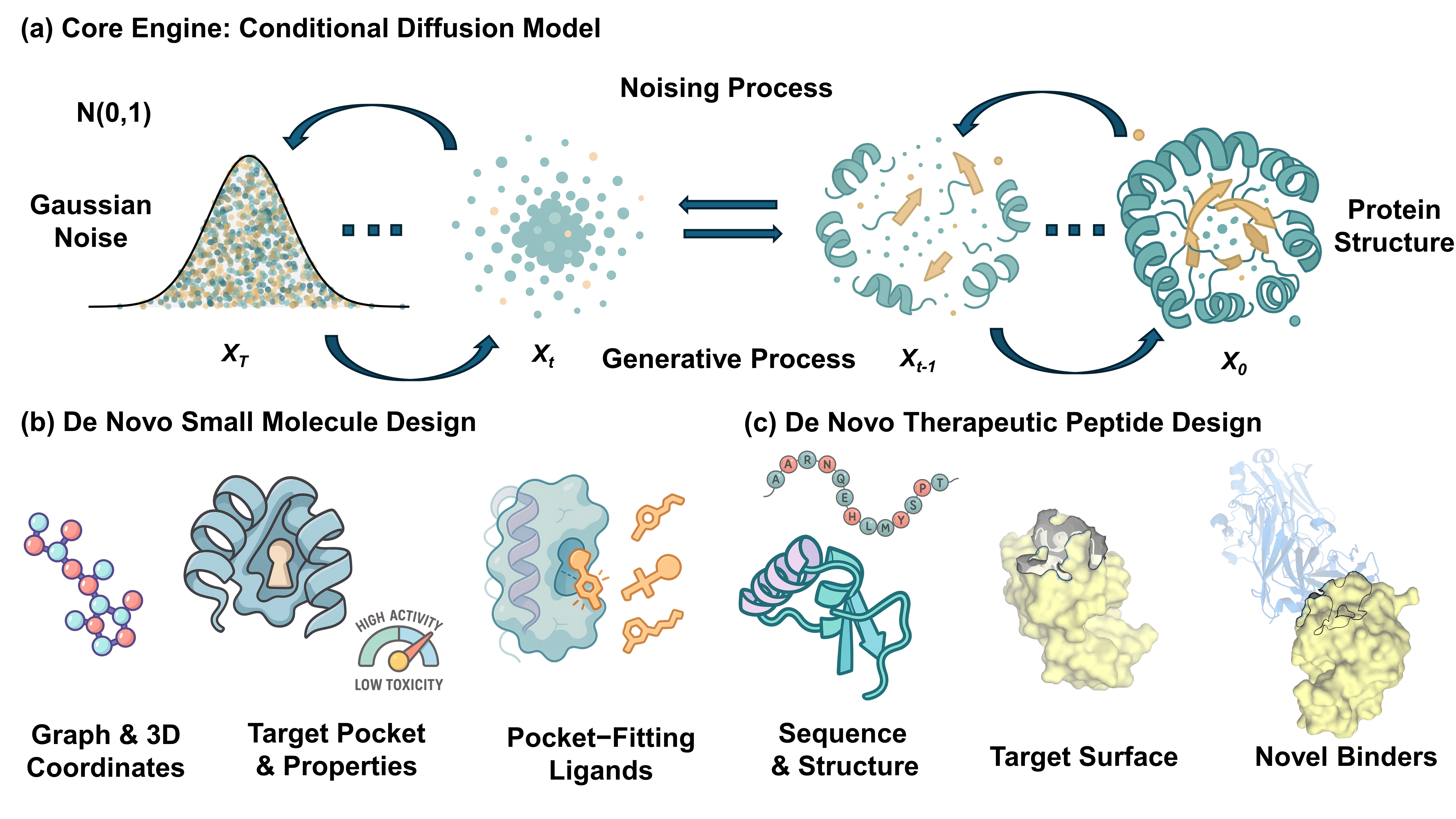
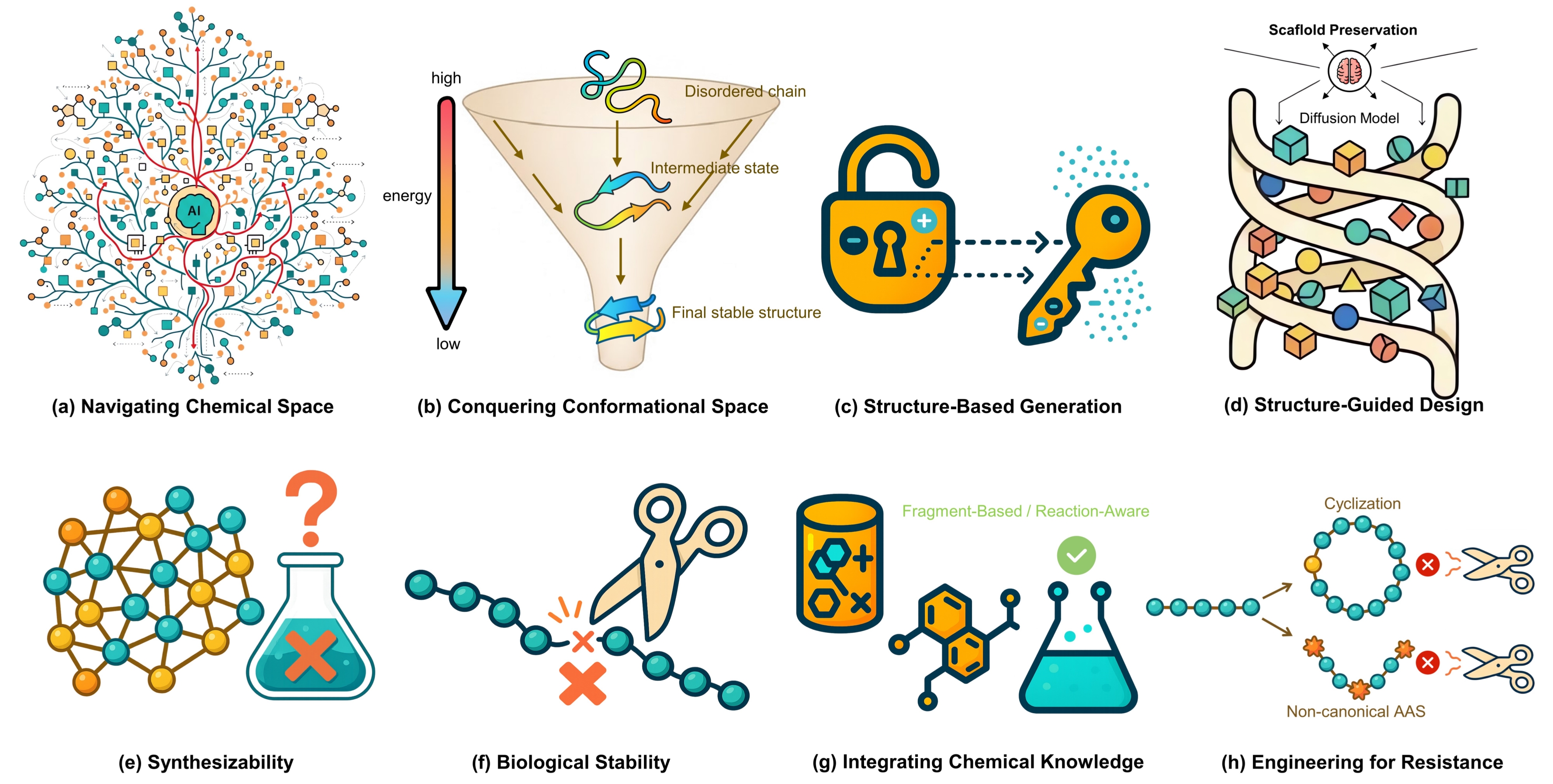
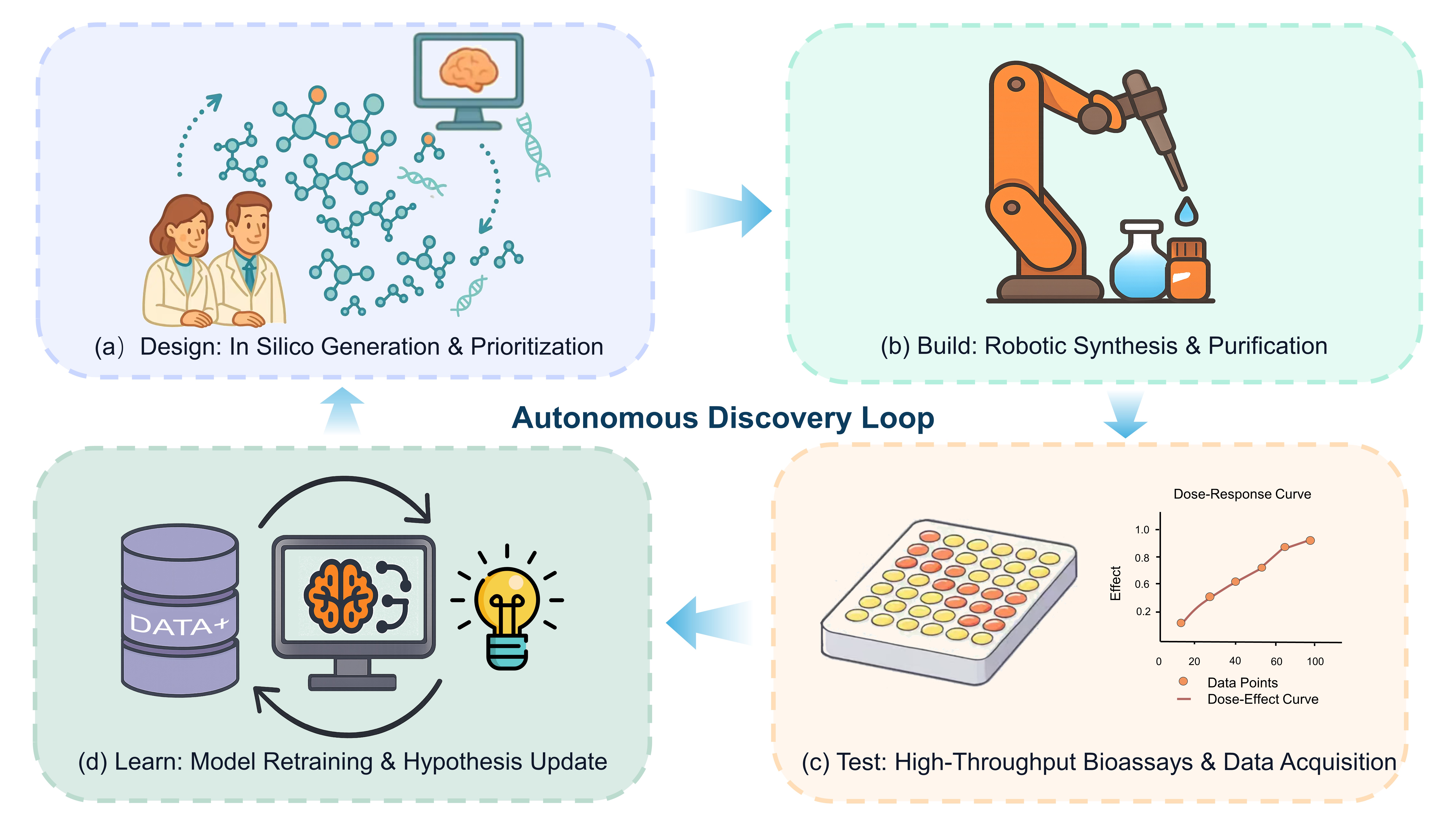
Diffusion models have emerged as a leading framework in generative modeling, poised to transform the traditionally slow and costly process of drug discovery. This review provides a systematic comparison of their application in designing two principal therapeutic modalities: small molecules and therapeutic peptides. We dissect how the unified framework of iterative denoising is adapted to the distinct molecular representations, chemical spaces, and design objectives of each modality. For small molecules, these models excel at structure-based design, generating novel, pocket-fitting ligands with desired physicochemical properties, yet face the critical hurdle of ensuring chemical synthesizability. Conversely, for therapeutic peptides, the focus shifts to generating functional sequences and designing de novo structures, where the primary challenges are achieving biological stability against proteolysis, ensuring proper folding, and minimizing immunogenicity. Despite these distinct challenges, both domains face shared hurdles: the scarcity of high-quality experimental data, the reliance on inaccurate scoring functions for validation, and the crucial need for experimental validation. We conclude that the full potential of diffusion models will be unlocked by bridging these modality-specific gaps and integrating them into automated, closed-loop Design-Build-Test-Learn (DBTL) platforms, thereby shifting the paradigm from mere chemical exploration to the on-demand engineering of novel~therapeutics.💡 Deep Analysis
📄 Full Content
📸 Image Gallery

Reference
This content is AI-processed based on open access ArXiv data.